SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies
- PMID: 32818852
- PMCID: PMC7418792
- DOI: 10.1016/j.jcv.2020.104585
SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies
Abstract
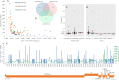
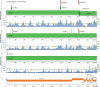
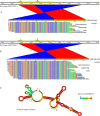
In December 2019, an outbreak of atypical pneumonia (Coronavirus disease 2019 -COVID-19) associated with a novel coronavirus (SARS-CoV-2) was reported in Wuhan city, Hubei province, China. The outbreak was traced to a seafood wholesale market and human to human transmission was confirmed. The rapid spread and the death toll of the new epidemic warrants immediate intervention. The intra-host genomic variability of SARS-CoV-2 plays a pivotal role in the development of effective antiviral agents and vaccines, as well as in the design of accurate diagnostics. We analyzed NGS data derived from clinical samples of three Chinese patients infected with SARS-CoV-2, in order to identify small- and large-scale intra-host variations in the viral genome. We identified tens of low- or higher- frequency single nucleotide variations (SNVs) with variable density across the viral genome, affecting 7 out of 10 protein-coding viral genes. The majority of these SNVs (72/104) corresponded to missense changes. The annotation of the identified SNVs but also of all currently circulating strain variations revealed colocalization of intra-host as well as strain specific SNVs with primers and probes currently used in molecular diagnostics assays. Moreover, we de-novo assembled the viral genome, in order to isolate and validate intra-host structural variations and recombination breakpoints. The bioinformatics analysis disclosed genomic rearrangements over poly-A / poly-U regions located in ORF1ab and spike (S) gene, including a potential recombination hot-spot within S gene. Our results highlight the intra-host genomic diversity and plasticity of SARS-CoV-2, pointing out genomic regions that are prone to alterations. The isolated SNVs and genomic rearrangements reflect the intra-patient capacity of the polymorphic quasispecies, which may arise rapidly during the outbreak, allowing immunological escape of the virus, offering resistance to anti-viral drugs and affecting the sensitivity of the molecular diagnostics assays.
Keywords: COVID-19 epidemic; Genomic rearrangements; Intra-host variability; Molecular diagnostics; Quasispecies; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Figures

References
-
- Fehr A.R., Perlman S. 2016. HHS Public Access; pp. 1–23. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

