Using spatial genetics to quantify mosquito dispersal for control programs
- PMID: 32819378
- PMCID: PMC7439557
- DOI: 10.1186/s12915-020-00841-0
Using spatial genetics to quantify mosquito dispersal for control programs
Abstract
Background: Hundreds of millions of people get a mosquito-borne disease every year and nearly one million die. Transmission of these infections is primarily tackled through the control of mosquito vectors. The accurate quantification of mosquito dispersal is critical for the design and optimization of vector control programs, yet the measurement of dispersal using traditional mark-release-recapture (MRR) methods is logistically challenging and often unrepresentative of an insect's true behavior. Using Aedes aegypti (a major arboviral vector) as a model and two study sites in Singapore, we show how mosquito dispersal can be characterized by the spatial analyses of genetic relatedness among individuals sampled over a short time span without interruption of their natural behaviors.
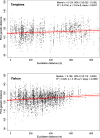
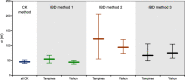
Results: Using simple oviposition traps, we captured adult female Ae. aegypti across high-rise apartment blocks and genotyped them using genome-wide SNP markers. We developed a methodology that produces a dispersal kernel for distance which results from one generation of successful breeding (effective dispersal), using the distance separating full siblings and 2nd- and 3rd-degree relatives (close kin). The estimated dispersal distance kernel was exponential (Laplacian), with a mean dispersal distance (and dispersal kernel spread σ) of 45.2 m (95% CI 39.7-51.3 m), and 10% probability of a dispersal > 100 m (95% CI 92-117 m). Our genetically derived estimates matched the parametrized dispersal kernels from previous MRR experiments. If few close kin are captured, a conventional genetic isolation-by-distance analysis can be used, as it can produce σ estimates congruent with the close-kin method if effective population density is accurately estimated. Genetic patch size, estimated by spatial autocorrelation analysis, reflects the spatial extent of the dispersal kernel "tail" that influences, for example, the critical radii of release zones and the speed of Wolbachia spread in mosquito replacement programs.
Conclusions: We demonstrate that spatial genetics can provide a robust characterization of mosquito dispersal. With the decreasing cost of next-generation sequencing, the production of spatial genetic data is increasingly accessible. Given the challenges of conventional MRR methods, and the importance of quantified dispersal in operational vector control decisions, we recommend genetic-based dispersal characterization as the more desirable means of parameterization.
Keywords: Close kin; Dispersal kernel; Genome-wide SNPs; IBD; Mosquito dispersal; Spatial autocorrelation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- World Health Assembly. WHO | Global vector control response 2017–2030: World Health Organization; 2018. http://www.who.int/vector-control/publications/global-control-response/en/. Accessed 10 Feb 2020.
-
- Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–457. - PubMed
-
- Kittayapong P, Kaeothaisong N-O, Ninphanomchai S, Limohpasmanee W. Combined sterile insect technique and incompatible insect technique: sex separation and quality of sterile Aedes aegypti male mosquitoes released in a pilot population suppression trial in Thailand. Parasit Vectors. 2018;11:657. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

