GRL-0920, an Indole Chloropyridinyl Ester, Completely Blocks SARS-CoV-2 Infection
- PMID: 32820005
- PMCID: PMC7441487
- DOI: 10.1128/mBio.01833-20
GRL-0920, an Indole Chloropyridinyl Ester, Completely Blocks SARS-CoV-2 Infection
Abstract
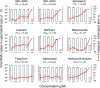
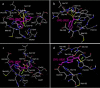
We assessed various newly generated compounds that target the main protease (Mpro) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and various previously known compounds reportedly active against SARS-CoV-2, employing RNA quantitative PCR (RNA-qPCR), cytopathicity assays, and immunocytochemistry. Here, we show that two indole-chloropyridinyl-ester derivatives, GRL-0820 and GRL-0920, exerted potent activity against SARS-CoV-2 in cell-based assays performed using VeroE6 cells and TMPRSS2-overexpressing VeroE6 cells. While GRL-0820 and the nucleotide analog remdesivir blocked SARS-CoV-2 infection, viral breakthrough occurred. No significant anti-SARS-CoV-2 activity was found for several compounds reportedly active against SARS-CoV-2 such as lopinavir, nelfinavir, nitazoxanide, favipiravir, and hydroxychroloquine. In contrast, GRL-0920 exerted potent activity against SARS-CoV-2 (50% effective concentration [EC50] = 2.8 μM) and dramatically reduced the infectivity, replication, and cytopathic effect of SARS-CoV-2 without significant toxicity as examined with immunocytochemistry. Structural modeling shows that indole and chloropyridinyl of the derivatives interact with two catalytic dyad residues of Mpro, Cys145 and His41, resulting in covalent bonding, which was verified using high-performance liquid chromatography-mass spectrometry (HPLC/MS), suggesting that the indole moiety is critical for the anti-SARS-CoV-2 activity of the derivatives. GRL-0920 might serve as a potential therapeutic for coronavirus disease 2019 (COVID-19) and might be optimized to generate more-potent anti-SARS-CoV-2 compounds.IMPORTANCE Targeting the main protease (Mpro) of SARS-CoV-2, we identified two indole-chloropyridinyl-ester derivatives, GRL-0820 and GRL-0920, active against SARS-CoV-2, employing RNA-qPCR and immunocytochemistry and show that the two compounds exerted potent activity against SARS-CoV-2. While GRL-0820 and remdesivir blocked SARS-CoV-2 infection, viral breakthrough occurred as examined with immunocytochemistry. In contrast, GRL-0920 completely blocked the infectivity and cytopathic effect of SARS-CoV-2 without significant toxicity. Structural modeling showed that indole and chloropyridinyl of the derivatives interacted with two catalytic dyad residues of Mpro, Cys145 and His41, resulting in covalent bonding, which was verified using HPLC/MS. The present data should shed light on the development of therapeutics for COVID-19, and optimization of GRL-0920 based on the present data is essential to develop more-potent anti-SARS-CoV-2 compounds for treating COVID-19.
Keywords: COVID-19; SARS-CoV-2; antiviral agents; main protease.
Figures



Comment in
-
Antiviral Drug Discovery To Address the COVID-19 Pandemic.mBio. 2020 Sep 25;11(5):e02134-20. doi: 10.1128/mBio.02134-20. mBio. 2020. PMID: 32978312 Free PMC article.
Similar articles
-
Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.Nat Commun. 2020 Sep 4;11(1):4417. doi: 10.1038/s41467-020-18233-x. Nat Commun. 2020. PMID: 32887884 Free PMC article.
-
Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy.ACS Comb Sci. 2020 Jun 8;22(6):297-305. doi: 10.1021/acscombsci.0c00058. Epub 2020 May 27. ACS Comb Sci. 2020. PMID: 32402186 Review.
-
Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.Nat Struct Mol Biol. 2020 Jun;27(6):529-532. doi: 10.1038/s41594-020-0440-6. Epub 2020 May 7. Nat Struct Mol Biol. 2020. PMID: 32382072
-
Structural-based virtual screening and in vitro assays for small molecules inhibiting the feline coronavirus 3CL protease as a surrogate platform for coronaviruses.Antiviral Res. 2020 Oct;182:104927. doi: 10.1016/j.antiviral.2020.104927. Epub 2020 Sep 7. Antiviral Res. 2020. PMID: 32910955 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
Cited by
-
Drug repurposing against SARS-CoV-1, SARS-CoV-2 and MERS-CoV.Future Microbiol. 2021 Nov;16:1341-1370. doi: 10.2217/fmb-2021-0019. Epub 2021 Nov 10. Future Microbiol. 2021. PMID: 34755538 Free PMC article.
-
Regulation of the Dimerization and Activity of SARS-CoV-2 Main Protease through Reversible Glutathionylation of Cysteine 300.mBio. 2021 Aug 31;12(4):e0209421. doi: 10.1128/mBio.02094-21. Epub 2021 Aug 17. mBio. 2021. PMID: 34399606 Free PMC article.
-
Detecting SARS-CoV-2 3CLpro expression and activity using a polyclonal antiserum and a luciferase-based biosensor.Virology. 2021 Apr;556:73-78. doi: 10.1016/j.virol.2021.01.010. Epub 2021 Jan 26. Virology. 2021. PMID: 33548599 Free PMC article.
-
Highly Neutralizing COVID-19 Convalescent Plasmas Potently Block SARS-CoV-2 Replication and Pneumonia in Syrian Hamsters.J Virol. 2022 Feb 23;96(4):e0155121. doi: 10.1128/JVI.01551-21. Epub 2021 Nov 24. J Virol. 2022. PMID: 34818068 Free PMC article.
-
Correlates of neutralizing/SARS-CoV-2-S1-binding antibody response with adverse effects and immune kinetics in BNT162b2-vaccinated individuals.Sci Rep. 2021 Nov 24;11(1):22848. doi: 10.1038/s41598-021-01930-y. Sci Rep. 2021. PMID: 34819514 Free PMC article.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. doi:10.1056/NEJMoa2001316. - DOI - PMC - PubMed
-
- World Health Organization. 2020. Coronavirus disease (COVID-19) situation report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 30 June 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
