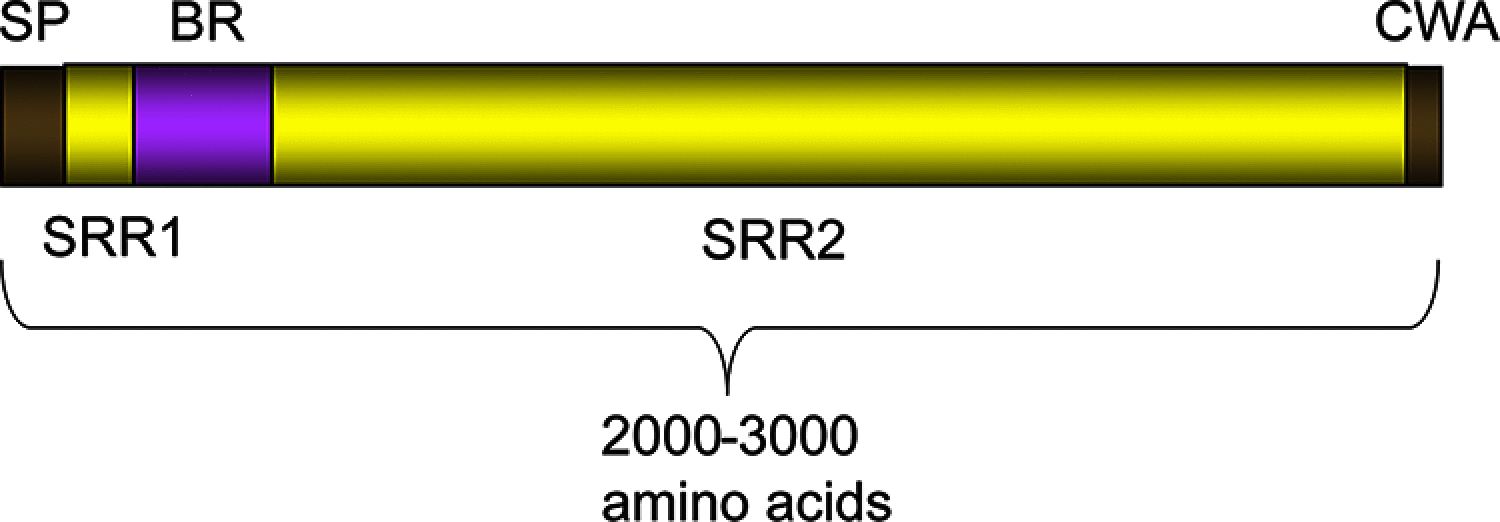
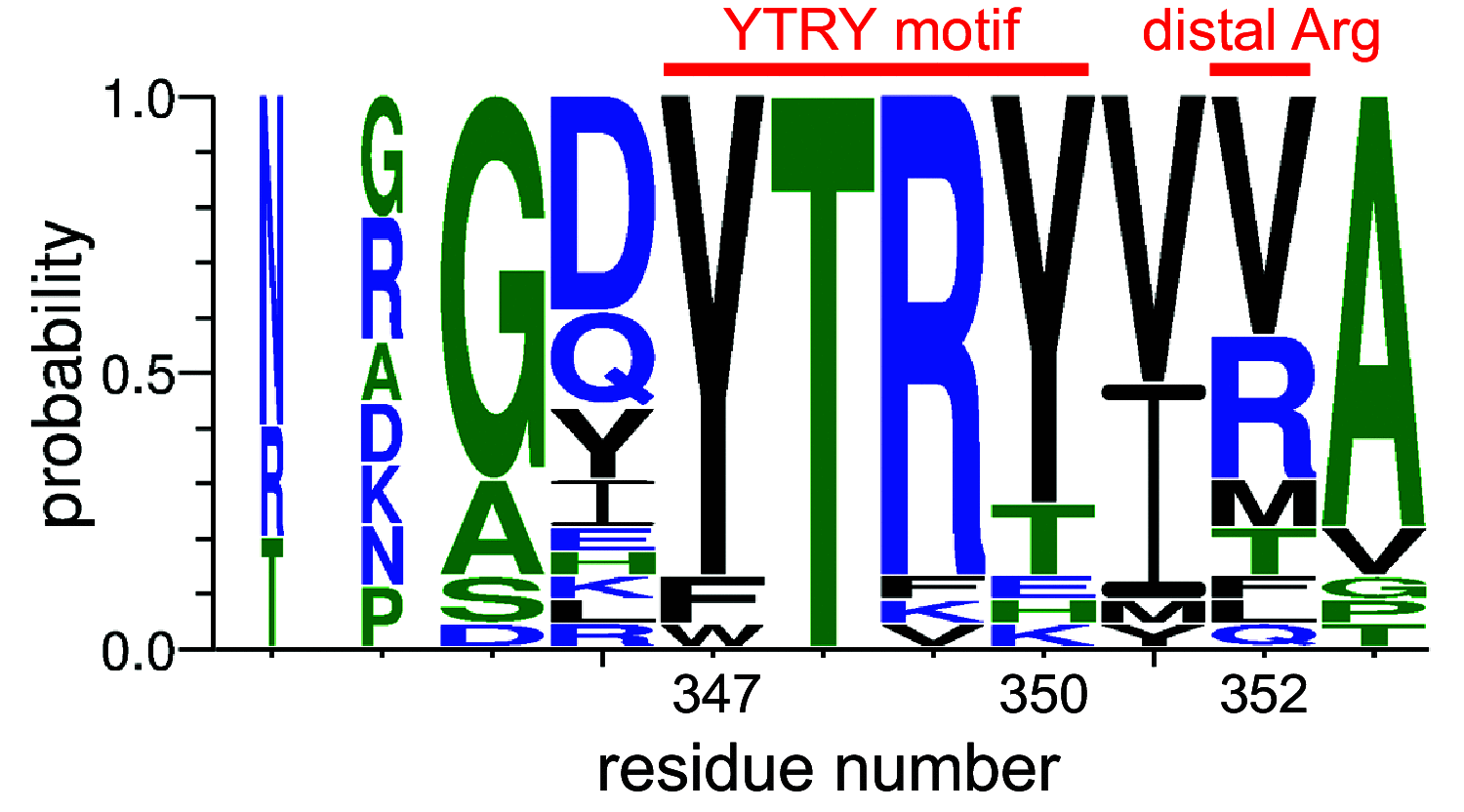
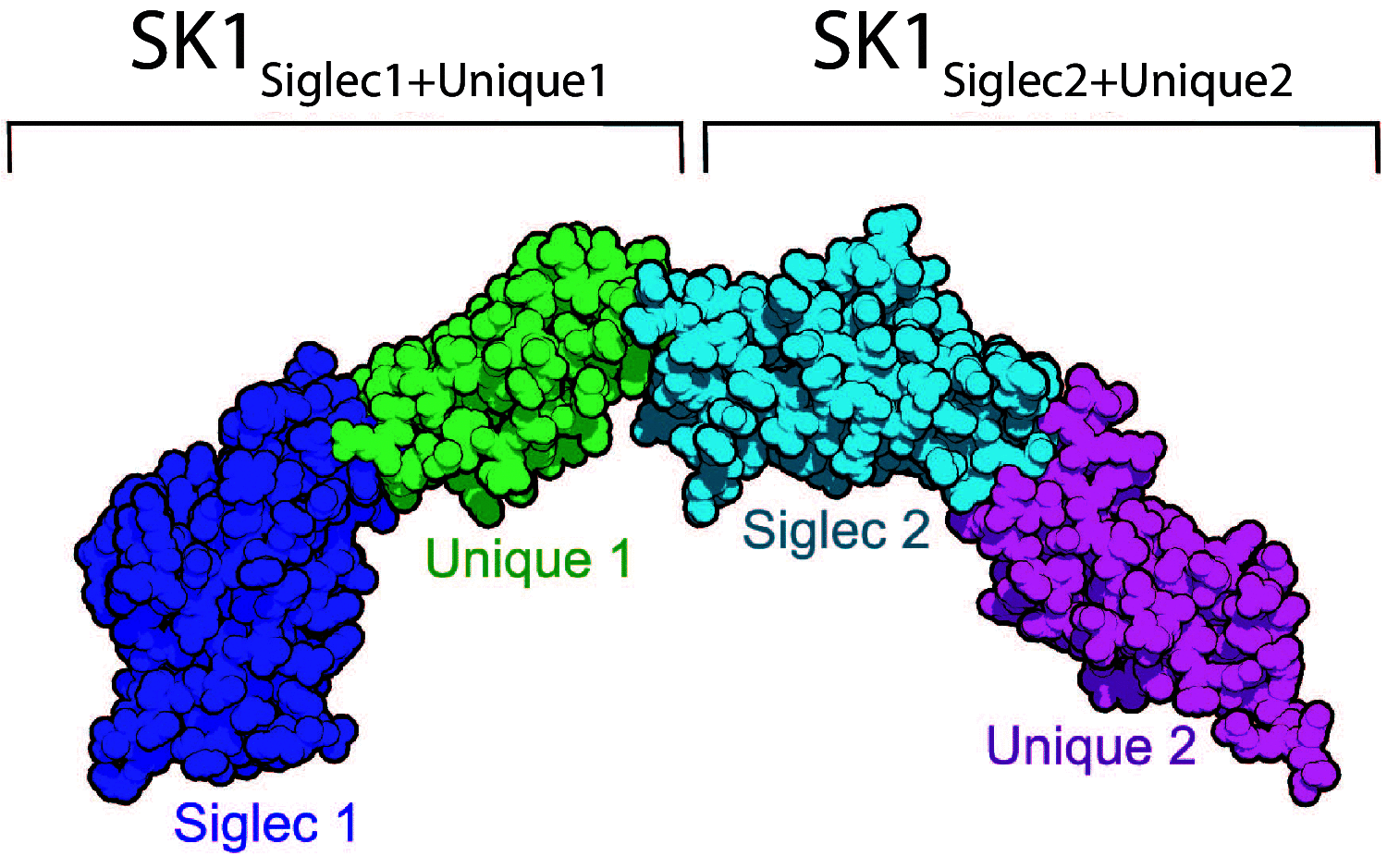
Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects
- PMID: 32820052
- PMCID: PMC7586212
- DOI: 10.1074/jbc.RA120.014177
Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects
Abstract
Keywords: Streptococcus; X-ray crystallography; adhesin; bacterial adhesion; bacterial pathogenesis; carbohydrate-binding protein; crystal structure; host-pathogen interaction; infectious disease; protein crystallization.
© 2020 Stubbs et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


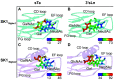

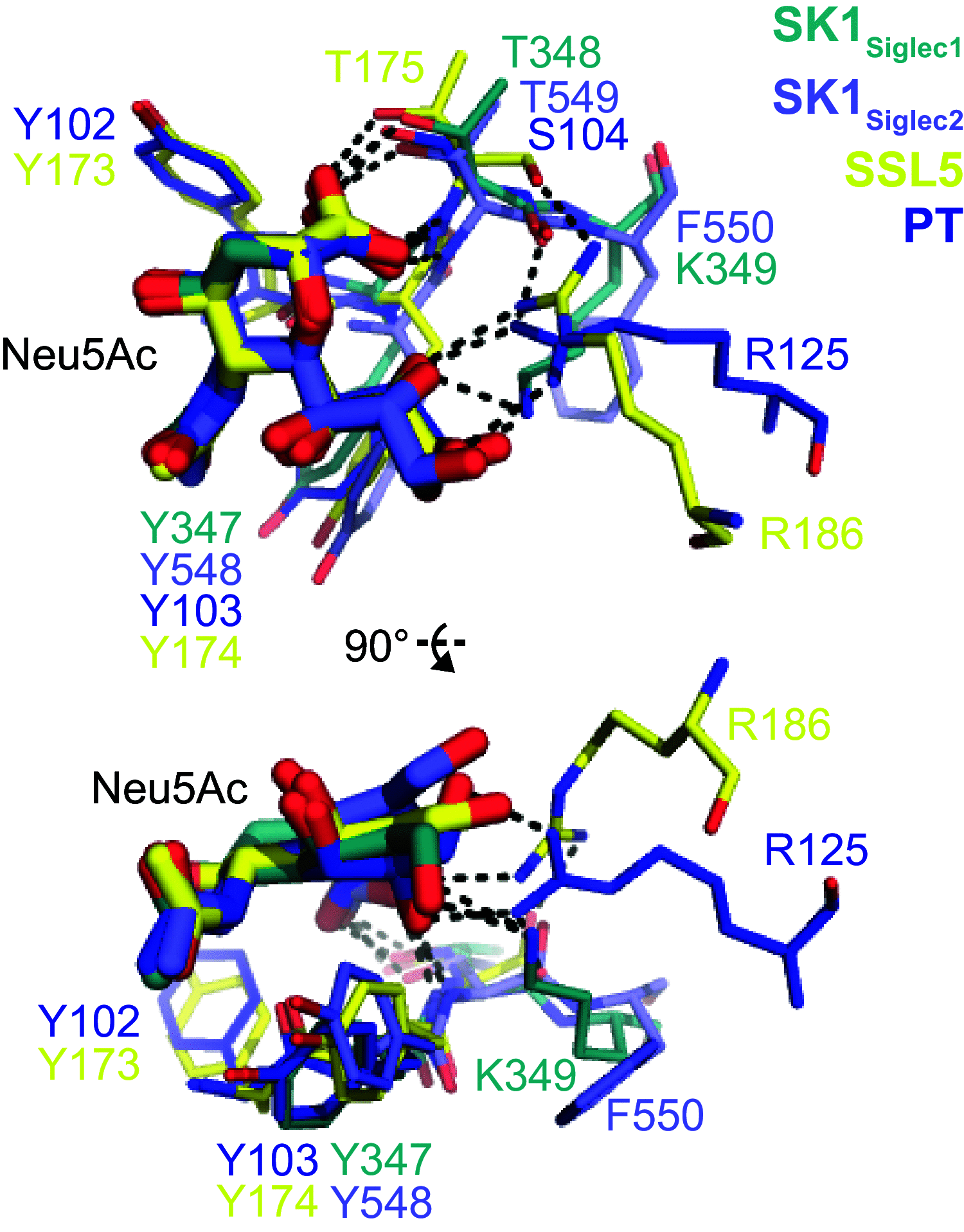


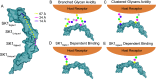
References
-
- Ramboarina S., Garnett J. A., Zhou M., Li Y., Peng Z., Taylor J. D., Lee W. C., Bodey A., Murray J. W., Alguel Y., Bergeron J., Bardiaux B., Sawyer E., Isaacson R., Tagliaferri C., et al. (2010) Structural insights into serine-rich fimbriae from Gram-positive bacteria. J. Biol. Chem. 285, 32446–32457 10.1074/jbc.M110.128165 - DOI - PMC - PubMed
-
- Deng L., Bensing B. A., Thamadilok S., Yu H., Lau K., Chen X., Ruhl S., Sullam P. M., and Varki A. (2014) Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 10, e1004540 10.1371/journal.ppat.1004540 - DOI - PMC - PubMed
-
- Ronis A., Brockman K., Singh A. K., Gaytán M. O., Wong A., McGrath S., Owen C. D., Magrini V., Wilson R. K., van der Linden M., and King S. J. (2019) Streptococcus oralis subsp. dentisani produces monolateral serine-rich repeat protein fibrils, one of which contributes to saliva binding via sialic acid. Infect. Immun. 87, e00406–19 10.1128/IAI.00406-19 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions

