Unpuzzling COVID-19: tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission
- PMID: 32820801
- PMCID: PMC7443512
- DOI: 10.1042/CS20200904
Unpuzzling COVID-19: tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission
Erratum in
-
Correction: Unpuzzling COVID-19: tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission.Clin Sci (Lond). 2020 Sep 18;134(17):2315-2317. doi: 10.1042/CS-20200904_COR. Clin Sci (Lond). 2020. PMID: 32901820 Free PMC article. No abstract available.
Abstract
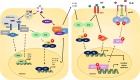
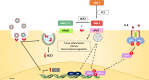
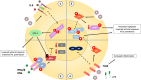
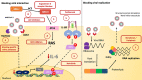
The highly infective coronavirus disease 19 (COVID-19) is caused by a novel strain of coronaviruses - the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) - discovered in December 2019 in the city of Wuhan (Hubei Province, China). Remarkably, COVID-19 has rapidly spread across all continents and turned into a public health emergency, which was ultimately declared as a pandemic by the World Health Organization (WHO) in early 2020. SARS-CoV-2 presents similar aspects to other members of the coronavirus family, mainly regarding its genome, protein structure and intracellular mechanisms, that may translate into mild (or even asymptomatic) to severe infectious conditions. Although the mechanistic features underlying the COVID-19 progression have not been fully clarified, current evidence have suggested that SARS-CoV-2 may primarily behave as other β-coronavirus members. To better understand the development and transmission of COVID-19, unveiling the signaling pathways that may be impacted by SARS-CoV-2 infection, at the molecular and cellular levels, is of crucial importance. In this review, we present the main aspects related to the origin, classification, etiology and clinical impact of SARS-CoV-2. Specifically, here we describe the potential mechanisms of cellular interaction and signaling pathways, elicited by functional receptors, in major targeted tissues/organs from the respiratory, gastrointestinal (GI), cardiovascular, renal, and nervous systems. Furthermore, the potential involvement of these signaling pathways in evoking the onset and progression of COVID-19 symptoms in these organ systems are presently discussed. A brief description of future perspectives related to potential COVID-19 treatments is also highlighted.
Keywords: COVID-19; Coronavirus; SARS-CoV-2; signaling pathway.
© 2020 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interest associated with the manuscript.
Figures

References
-
- Almeida J.D., Berry D.M., Cunningham C.H., Hamre D., Hofstad M.S., Mallucci L. et al. . (1968) Coronaviruses. Nature 220, 2
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

