Senotherapeutic drugs for human intervertebral disc degeneration and low back pain
- PMID: 32821059
- PMCID: PMC7442487
- DOI: 10.7554/eLife.54693
Senotherapeutic drugs for human intervertebral disc degeneration and low back pain
Abstract
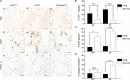
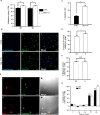
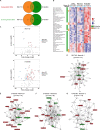
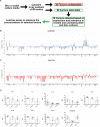
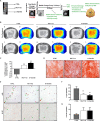
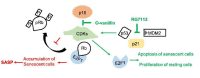
Cellular senescence is a contributor to intervertebral disc (IVD) degeneration and low back pain. Here, we found that RG-7112, a potent mouse double-minute two protein inhibitor, selectively kills senescent IVD cells through apoptosis. Gene expression pathway analysis was used to compare the functional networks of genes affected by RG-7112, a pure synthetic senolytic with o-Vanillin a natural and anti-inflammatory senolytic. Both affected a functional gene network related to cell death and survival. O-Vanillin also affected networks related to cell cycle progression as well as connective tissue development and function. Both senolytics effectively decreased the senescence-associated secretory phenotype (SASP) of IVD cells. Furthermore, bioavailability and efficacy were verified ex vivo in the physiological environment of degenerating intact human discs where a single dose improved disc matrix homeostasis. Matrix improvement correlated with a reduction in senescent cells and SASP, supporting a translational potential of targeting senescent cells as a therapeutic intervention.
Keywords: annulus fibrosus; cartilage; cell biology; human; intervertebral disc; medicine; nucleus pulposus.
Plain language summary
Pain in the lower back affects about four in five people during their lifetime. Over time, the discs that provide cushioning between the vertebrae of the spine can degenerate, which can be one of the major causes of lower back pain. It has been shown that when the cells of these discs are exposed to different stress factors, they stop growing and become irreversibly dormant. Such ‘senescent’ cells release a range of proteins and small molecules that lead to painful inflammation and further degeneration of the discs. Moreover, it is thought that a high number of senescent cells may be linked to other degenerative diseases such as arthritis. Current treatments can only reduce the severity of the symptoms, but they cannot prevent the degeneration from progressing. Now, Cherif et al. set out to test the effects of two different compounds on human disc cells grown in the laboratory. One of the molecules studied, RG-7112, is a synthetic drug that has been approved for safety by the US Food and Drug Administration and has been shown to remove senescent cells. The other, o-Vanillin, is a natural compound that has anti-inflammatory and anti-senescence properties. The results showed that both compounds were able to trigger changes to that helped new, healthy cells to grow and at the same time kill senescent cells. They also reduced the production of molecules linked to inflammation and pain. Further analyses revealed that the compounds were able to strengthen the fibrous matrix that surrounds and supports the discs. Cherif et al. hope that this could form the basis for a new family of drugs for back pain to slow the degeneration of the discs and reduce pain. This may also have benefits for other similar degenerative diseases caused by cell senescence, such as arthritis.
© 2020, Cherif et al.
Conflict of interest statement
HC, DB, MM, OR, JO, LH No competing interests declared
Figures




References
-
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, d'Adda di Fagagna F, Bernard D, Hernando E, Gil J. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. - DOI - PubMed
-
- Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang TW, Lasitschka F, Andrulis M, Pascual G, Morris KJ, Khan S, Jin H, Dharmalingam G, Snijders AP, Carroll T, Capper D, Pritchard C, Inman GJ, Longerich T, Sansom OJ, Benitah SA, Zender L, Gil J. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nature Cell Biology. 2013;15:978–990. doi: 10.1038/ncb2784. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

