The Polycomb-associated factor PHF19 controls hematopoietic stem cell state and differentiation
- PMID: 32821835
- PMCID: PMC7406347
- DOI: 10.1126/sciadv.abb2745
The Polycomb-associated factor PHF19 controls hematopoietic stem cell state and differentiation
Abstract
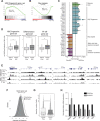
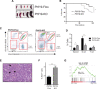
Adult hematopoietic stem cells (HSCs) are rare multipotent cells in bone marrow that are responsible for generating all blood cell types. HSCs are a heterogeneous group of cells with high plasticity, in part, conferred by epigenetic mechanisms. PHF19, a subunit of the Polycomb repressive complex 2 (PRC2), is preferentially expressed in mouse hematopoietic precursors. Here, we now show that, in stark contrast to results published for other PRC2 subunits, genetic depletion of Phf19 increases HSC identity and quiescence. While proliferation of HSCs is normally triggered by forced mobilization, defects in differentiation impede long-term correct blood production, eventually leading to aberrant hematopoiesis. At molecular level, PHF19 deletion triggers a redistribution of the histone repressive mark H3K27me3, which notably accumulates at blood lineage-specific genes. Our results provide novel insights into how epigenetic mechanisms determine HSC identity, control differentiation, and are key for proper hematopoiesis.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures


References
-
- Haas S., Trumpp A., Milsom M. D., Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 22, 627–638 (2018). - PubMed
-
- Plass C., Pfister S. M., Lindroth A. M., Bogatyrova O., Claus R., Lichter P., Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet. 14, 765–780 (2013). - PubMed
-
- Di Croce L., Helin K., Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155 (2013). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

