Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo
- PMID: 32821836
- PMCID: PMC7413728
- DOI: 10.1126/sciadv.abb3521
Positive dielectrophoresis-based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo
Abstract
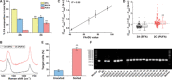
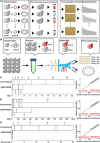
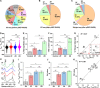
The potential of Raman-activated cell sorting (RACS) is inherently limited by conflicting demands for signal quality and sorting throughput. Here, we present positive dielectrophoresis-based Raman-activated droplet sorting (pDEP-RADS), where a periodical pDEP force was exerted to trap fast-moving cells, followed by simultaneous microdroplet encapsulation and sorting. Screening of yeasts for triacylglycerol (TAG) content demonstrated near-theoretical-limit accuracy, ~120 cells min-1 throughput and full-vitality preservation, while sorting fatty acid degree of unsaturation (FA-DU) featured ~82% accuracy at ~40 cells min-1. From a yeast library expressing algal diacylglycerol acyltransferases (DGATs), a pDEP-RADS run revealed all reported TAG-synthetic variants and distinguished FA-DUs of enzyme products. Furthermore, two previously unknown DGATs producing low levels of monounsaturated fatty acid-rich TAG were discovered. This first demonstration of RACS for enzyme discovery represents hundred-fold saving in time consumables and labor versus culture-based approaches. The ability to automatically flow-sort resonance Raman-independent phenotypes greatly expands RACS' application.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- He Y., Wang X., Ma B., Xu J., Ramanome technology platform for label-free screening and sorting of microbial cell factories at single-cell resolution. Biotechnol. Adv. 37, 107388 (2019). - PubMed
-
- Song Y., Yin H., Huang W. E., Raman activated cell sorting. Curr. Opin. Chem. Biol. 33, 1–8 (2016). - PubMed
-
- Zhang Q., Zhang P., Gou H., Mou C., Huang W. E., Yang M., Xu J., Ma B., Towards high-throughput microfluidic Raman-activated cell sorting. Analyst 140, 6163–6174 (2015). - PubMed
-
- Huang W. E., Ward A. D., Whiteley A. S., Raman tweezers sorting of single microbial cells. Environ. Microbiol. Rep. 1, 44–49 (2009). - PubMed
-
- Berry D., Mader E., Lee T. K., Woebken D., Wang Y., Zhu D., Palatinszky M., Schintlmeister A., Schmid M. C., Hanson B. T., Shterzer N., Mizrahi I., Rauch I., Decker T., Bocklitz T., Popp J., Gibson C. M., Fowler P. W., Huang W. E., Wagner M., Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 112, E194–E203 (2015). - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

