Decidual NK Cells Transfer Granulysin to Selectively Kill Bacteria in Trophoblasts
- PMID: 32822574
- PMCID: PMC7484179
- DOI: 10.1016/j.cell.2020.07.019
Decidual NK Cells Transfer Granulysin to Selectively Kill Bacteria in Trophoblasts
Abstract
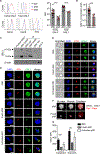
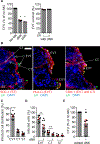
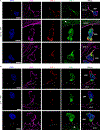
Maternal decidual NK (dNK) cells promote placentation, but how they protect against placental infection while maintaining fetal tolerance is unclear. Here we show that human dNK cells highly express the antimicrobial peptide granulysin (GNLY) and selectively transfer it via nanotubes to extravillous trophoblasts to kill intracellular Listeria monocytogenes (Lm) without killing the trophoblast. Transfer of GNLY, but not other cell death-inducing cytotoxic granule proteins, strongly inhibits Lm in human placental cultures and in mouse and human trophoblast cell lines. Placental and fetal Lm loads are lower and pregnancy success is greatly improved in pregnant Lm-infected GNLY-transgenic mice than in wild-type mice that lack GNLY. This immune defense is not restricted to pregnancy; peripheral NK (pNK) cells also transfer GNLY to kill bacteria in macrophages and dendritic cells without killing the host cell. Nanotube transfer of GNLY allows dNK to protect against infection while leaving the maternal-fetal barrier intact.
Keywords: Listeria monocytogenes; cytotoxic granule; decidual NK cells; extravillous trophoblast; granulysin; maternal-fetal tolerance; nanotube; pregnancy.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
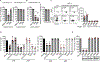


Comment in
-
Killing via nanotubes.Nat Rev Immunol. 2020 Oct;20(10):592-593. doi: 10.1038/s41577-020-00444-4. Nat Rev Immunol. 2020. PMID: 32873890 No abstract available.
-
Stealth Killing by Uterine NK Cells for Tolerance and Tissue Homeostasis.Cell. 2020 Sep 3;182(5):1074-1076. doi: 10.1016/j.cell.2020.08.018. Cell. 2020. PMID: 32888492
-
Killing the Pathogen and Sparing the Placenta.N Engl J Med. 2020 Nov 19;383(21):2080-2082. doi: 10.1056/NEJMcibr2028357. N Engl J Med. 2020. PMID: 33207100 No abstract available.
References
-
- Berges C, Naujokat C, Tinapp S, Wieczorek H, Ho A, Sadeghi M, Opelz G, and Daniel V (2005). A cell line model for the differentiation of human dendritic cells. Biochem. Biophys. Res. Commun 333, 896–907. - PubMed
-
- Błaszkowska J, and Góralska K (2014). Parasites and fungi as a threat for prenatal and postnatal human development. Ann. Parasitol 60, 225–234. - PubMed
-
- Cao B, and Mysorekar IU (2014). Intracellular bacteria in placental basal plate localize to extravillous trophoblasts. Placenta 35, 139–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

