Canonical Wnt Pathway Controls mESC Self-Renewal Through Inhibition of Spontaneous Differentiation via β-Catenin/TCF/LEF Functions
- PMID: 32822589
- PMCID: PMC7486219
- DOI: 10.1016/j.stemcr.2020.07.019
Canonical Wnt Pathway Controls mESC Self-Renewal Through Inhibition of Spontaneous Differentiation via β-Catenin/TCF/LEF Functions
Abstract
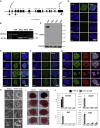
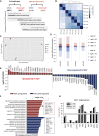
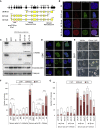
The Wnt/β-catenin signaling pathway is a key regulator of embryonic stem cell (ESC) self-renewal and differentiation. Constitutive activation of this pathway has been shown to increase mouse ESC (mESC) self-renewal and pluripotency gene expression. In this study, we generated a novel β-catenin knockout model in mESCs to delete putatively functional N-terminally truncated isoforms observed in previous knockout models. We showed that aberrant N-terminally truncated isoforms are not functional in mESCs. In the generated knockout line, we observed that canonical Wnt signaling is not active, as β-catenin ablation does not alter mESC transcriptional profile in serum/LIF culture conditions. In addition, we observed that Wnt signaling activation represses mESC spontaneous differentiation in a β-catenin-dependent manner. Finally, β-catenin (ΔC) isoforms can rescue β-catenin knockout self-renewal defects in mESCs cultured in serum-free medium and, albeit transcriptionally silent, cooperate with TCF1 and LEF1 to inhibit mESC spontaneous differentiation in a GSK3-dependent manner.
Keywords: CRISPR; Ctnnb1; LEF; TCF; Wnt; embryonic stem cells; mESCs; pluripotency; self-renewal; β-catenin.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Anton R., Kestler H.A., Kuhl M. Beta-catenin signaling contributes to stemness and regulates early differentiation in murine embryonic stem cells. FEBS Lett. 2007;581:5247–5254. - PubMed
-
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

