Adsorption Sequencing as a Rapid Method to Link Environmental Bacteriophages to Hosts
- PMID: 32823052
- PMCID: PMC7452251
- DOI: 10.1016/j.isci.2020.101439
Adsorption Sequencing as a Rapid Method to Link Environmental Bacteriophages to Hosts
Abstract
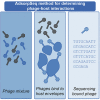
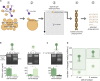
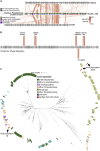
An important viromics challenge is associating bacteriophages to hosts. To address this, we developed adsorption sequencing (AdsorpSeq), a readily implementable method to measure phages that are preferentially adsorbed to specific host cell envelopes. AdsorpSeq thus captures the key initial infection cycle step. Phages are added to cell envelopes, adsorbed phages are isolated through gel electrophoresis, after which adsorbed phage DNA is sequenced and compared with the full virome. Here, we show that AdsorpSeq allows for separation of phages based on receptor-adsorbing capabilities. Next, we applied AdsorpSeq to identify phages in a wastewater virome that adsorb to cell envelopes of nine bacteria, including important pathogens. We detected 26 adsorbed phages including common and rare members of the virome, a minority being related to previously characterized phages. We conclude that AdsorpSeq is an effective new tool for rapid characterization of environmental phage adsorption, with a proof-of-principle application to Gram-negative host cell envelopes.
Keywords: Ecology; Environmental Science; Microbiology; Techniques in Genetics.
© 2020 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Abedon S.T., Yin J. Bacteriophage plaques: theory and analysis. In: Clokie M.R.J., Kropinski A., editors. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Humana Press; 2009. pp. 161–174.
LinkOut - more resources
Full Text Sources

