mTOR Inhibition Leads to Src-Mediated EGFR Internalisation and Degradation in Glioma Cells
- PMID: 32823532
- PMCID: PMC7464593
- DOI: 10.3390/cancers12082266
mTOR Inhibition Leads to Src-Mediated EGFR Internalisation and Degradation in Glioma Cells
Abstract
Epidermal Growth Factor receptor (EGFR) is a tyrosine kinase receptor widely expressed on the surface of numerous cell types, which activates several downstream signalling pathways involved in cell proliferation, migration and survival. EGFR alterations, such as overexpression or mutations, have been frequently observed in several cancers, including glioblastoma (GBM), and are associated to uncontrolled cell proliferation. Here we show that the inhibition of mammalian target of Rapamycin (mTOR) mediates EGFR delivery to lysosomes for degradation in GBM cells, independently of autophagy activation. Coherently with EGFR internalisation and degradation, mTOR blockade negatively affects the mitogen activated protein/extracellular signal-regulated kinase (MAPK)/ERK pathway. Furthermore, we provide evidence that Src kinase activation is required for EGFR internaliation upon mTOR inhibition. Our results further support the hypothesis that mTOR targeting may represent an effective therapeutic strategy in GBM management, as its inhibition results in EGFR degradation and in proliferative signal alteration.
Keywords: EGFR; autophagy; glioma; mTOR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
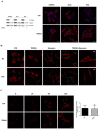
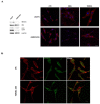
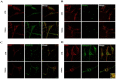


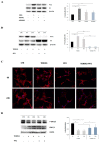
References
-
- Ceresa B.P., Peterson J.L. Cell and Molecular Biology of Epidermal Growth Factor Receptor. Int. Rev. Cell Mol. Biol. 2014;313:145–178. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

