The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles
- PMID: 32823751
- PMCID: PMC7463600
- DOI: 10.3390/cells9081898
The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles
Abstract
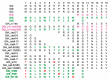
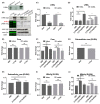
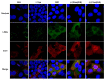
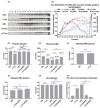
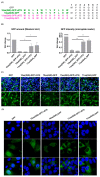
The N-terminus of the hepatitis B virus (HBV) large surface protein (LHB) differs with respect to genotypes. Compared to the amino terminus of genotype (Gt)D, in GtA, GtB and GtC, an additional identical 11 amino acids (aa) are found, while GtE and GtG share another similar 10 aa. Variants of GtB and GtC affecting this N-terminal part are associated with hepatoma formation. Deletion of these amino-terminal 11 aa in GtA reduces the amount of LHBs and changes subcellular accumulation (GtA-like pattern) to a dispersed distribution (GtD-like pattern). Vice versa, the fusion of the GtA-derived N-terminal 11 aa to GtD causes a GtA-like phenotype. However, insertion of the corresponding GtE-derived 10 aa to GtD has no effect. Deletion of these 11aa decreases filament size while neither the number of released viral genomes nor virion size and infectivity are affected. A negative regulatory element (aa 2-8) and a dominant positive regulatory element (aa 9-11) affecting the amount of LHBs were identified. The fusion of this motif to eGFP revealed that the effect on protein amount and subcellular distribution is not restricted to LHBs. These data identify a novel region in the N-terminus of LHBs affecting the amount and subcellular distribution of LHBs and identify release-promoting and -inhibiting aa residues within this motive.
Keywords: HBV filaments; HBV genotypes; HBV surface protein; PreS1 deletion; morphogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

