Molecular Epidemiology Analysis of SARS-CoV-2 Strains Circulating in Romania during the First Months of the Pandemic
- PMID: 32823907
- PMCID: PMC7460100
- DOI: 10.3390/life10080152
Molecular Epidemiology Analysis of SARS-CoV-2 Strains Circulating in Romania during the First Months of the Pandemic
Abstract
Background: The spread of SARS-CoV-2 generated an unprecedented global public health crisis. Soon after Asia, Europe was seriously affected. Many countries, including Romania, adopted lockdown measures to limit the outbreak.
Aim: We performed a molecular epidemiology analysis of SARS-CoV-2 viral strains circulating in Romania during the first two months of the epidemic in order to detect mutation profiles and phylogenetic relatedness.
Methods: Respiratory samples were directly used for shotgun sequencing.
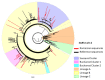
Results: All Romanian sequences belonged to lineage B, with a different subtype distribution between northern and southern regions (subtype B.1.5 and B.1.1). Phylogenetic analysis suggested that the Romanian epidemic started with multiple introduction events from other European countries followed by local transmission. Phylogenetic links between northern Romania and Spain, Austria, Scotland and Russia were observed, as well as between southern Romania and Switzerland, Italy, France and Turkey. One viral strain presented a previously unreported mutation in the Nsp2 gene, namely K489E. Epidemiologically-defined clusters displayed specific mutations, suggesting molecular signatures for strains coming from areas that were isolated during the lockdown.
Conclusions: Romanian epidemic was initiated by multiple introductions from European countries followed by local transmissions. Different subtype distribution between northern and southern Romania was observed after two months of the pandemic.
Keywords: Romania; SARS-CoV-2; WGS; coronavirus; mutations; outbreak; pandemic; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest. The sponsor “Mereu Aproape” Foundation had no role in the design, execution, interpretation, or writing of the study.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

