Critical Analysis of Genome-Wide Association Studies: Triple Negative Breast Cancer Quae Exempli Causa
- PMID: 32823908
- PMCID: PMC7461549
- DOI: 10.3390/ijms21165835
Critical Analysis of Genome-Wide Association Studies: Triple Negative Breast Cancer Quae Exempli Causa
Abstract
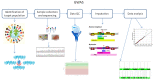
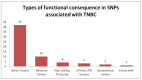
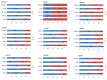
Genome-wide association studies (GWAS) are useful in assessing and analyzing either differences or variations in DNA sequences across the human genome to detect genetic risk factors of diseases prevalent within a target population under study. The ultimate goal of GWAS is to predict either disease risk or disease progression by identifying genetic risk factors. These risk factors will define the biological basis of disease susceptibility for the purposes of developing innovative, preventative, and therapeutic strategies. As single nucleotide polymorphisms (SNPs) are often used in GWAS, their relevance for triple negative breast cancer (TNBC) will be assessed in this review. Furthermore, as there are different levels and patterns of linkage disequilibrium (LD) present within different human subpopulations, a plausible strategy to evaluate known SNPs associated with incidence of breast cancer in ethnically different patient cohorts will be presented and discussed. Additionally, a description of GWAS for TNBC will be presented, involving various identified SNPs correlated with miRNA sites to determine their efficacies on either prognosis or progression of TNBC in patients. Although GWAS have identified multiple common breast cancer susceptibility variants that individually would result in minor risks, it is their combined effects that would likely result in major risks. Thus, one approach to quantify synergistic effects of such common variants is to utilize polygenic risk scores. Therefore, studies utilizing predictive risk scores (PRSs) based on known breast cancer susceptibility SNPs will be evaluated. Such PRSs are potentially useful in improving stratification for screening, particularly when combining family history, other risk factors, and risk prediction models. In conclusion, although interpretation of the results from GWAS remains a challenge, the use of SNPs associated with TNBC may elucidate and better contextualize these studies.
Keywords: CNVs; GWAS; SNPs; TNBC; breast cancer; linkage disequilibrium; predictive risk scores; structural variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

