MPN: The Molecular Drivers of Disease Initiation, Progression and Transformation and their Effect on Treatment
- PMID: 32823933
- PMCID: PMC7465511
- DOI: 10.3390/cells9081901
MPN: The Molecular Drivers of Disease Initiation, Progression and Transformation and their Effect on Treatment
Abstract
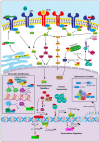
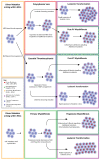
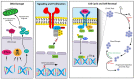
Myeloproliferative neoplasms (MPNs) constitute a group of disorders identified by an overproduction of cells derived from myeloid lineage. The majority of MPNs have an identifiable driver mutation responsible for cytokine-independent proliferative signalling. The acquisition of coexisting mutations in chromatin modifiers, spliceosome complex components, DNA methylation modifiers, tumour suppressors and transcriptional regulators have been identified as major pathways for disease progression and leukemic transformation. They also confer different sensitivities to therapeutic options. This review will explore the molecular basis of MPN pathogenesis and specifically examine the impact of coexisting mutations on disease biology and therapeutic options.
Keywords: CALR; DNA methylation; IFNα; JAK2; MPL; MPN; chromatin modifiers; driver mutations; leukemic transformation; myeloproliferation; spliceosome; transcriptional regulators; tumour suppressors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
-
- Tefferi A., Rumi E., Finazzi G., Gisslinger H., Vannucchi A.M., Rodeghiero F., Randi M.L., Vaidya R., Cazzola M., Rambaldi A., et al. Survival and prognosis among 1545 patients with contemporary polycythemia vera: An international study. Leukemia. 2013;27:1874–1881. doi: 10.1038/leu.2013.163. - DOI - PMC - PubMed
-
- Vannucchi A.M., Antonioli E., Guglielmelli P., Longo G., Pancrazzi A., Ponziani V., Bogani C., Ferrini P.R., Rambaldi A., Guerini V., et al. Prospective identification of high-risk polycythemia vera patients based on JAK2V617F allele burden. Leukemia. 2007;21:1952–1959. doi: 10.1038/sj.leu.2404854. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

