The Role of APOSTART in Switching between Sexuality and Apomixis in Poa pratensis
- PMID: 32824095
- PMCID: PMC7464379
- DOI: 10.3390/genes11080941
The Role of APOSTART in Switching between Sexuality and Apomixis in Poa pratensis
Abstract
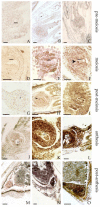
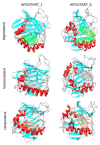
The production of seeds without sex is considered the holy grail of plant biology. The transfer of apomixis to various crop species has the potential to transform plant breeding, since it will allow new varieties to retain valuable traits thorough asexual reproduction. Therefore, a greater molecular understanding of apomixis is fundamental. In a previous work we identified a gene, namely APOSTART, that seemed to be involved in this asexual mode of reproduction, which is very common in Poa pratensis L., and here we present a detailed work aimed at clarifying its role in apomixis. In situ hybridization showed that PpAPOSTART is expressed in reproductive tissues from pre-meiosis to embryo development. Interestingly, it is expressed early in few nucellar cells of apomictic individuals possibly switching from a somatic to a reproductive cell as in aposporic apomixis. Moreover, out of 13 APOSTART members, we identified one, APOSTART_6, as specifically expressed in flower tissue. APOSTART_6 also exhibited delayed expression in apomictic genotypes when compared with sexual types. Most importantly, the SCAR (Sequence Characterized Amplified Region) derived from the APOSTART_6 sequence completely co-segregated with apomixis.
Keywords: APOSTART; Poa pratensis; apomixis; plant reproduction.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of manuscript or in the decision to publish the results.
Figures



References
-
- Bashaw E.C. Problems and possibilities of apomixis in the improvement of tropical forage grasses. Trop. Forages Livest. Prod. Syst. 1975;24:23–30.
-
- Hanna W., Bashaw E.C. Apomixis: Its identification and use in plant breeding 1. Crop Sci. 1987;27:1136–1139. doi: 10.2135/cropsci1987.0011183X002700060010x. - DOI
-
- Savidan Y., Dujardin M. Apomixie: La prochaine révolution verte? Rech. Paris 1970. 1992;23:326–334.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

