Progress in Developing Inhibitors of SARS-CoV-2 3C-Like Protease
- PMID: 32824639
- PMCID: PMC7463875
- DOI: 10.3390/microorganisms8081250
Progress in Developing Inhibitors of SARS-CoV-2 3C-Like Protease
Abstract
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The viral outbreak started in late 2019 and rapidly became a serious health threat to the global population. COVID-19 was declared a pandemic by the World Health Organization in March 2020. Several therapeutic options have been adopted to prevent the spread of the virus. Although vaccines have been developed, antivirals are still needed to combat the infection of this virus. SARS-CoV-2 is an enveloped virus, and its genome encodes polyproteins that can be processed into structural and nonstructural proteins. Maturation of viral proteins requires cleavages by proteases. Therefore, the main protease (3 chymotrypsin-like protease (3CLpro) or Mpro) encoded by the viral genome is an attractive drug target because it plays an important role in cleaving viral polyproteins into functional proteins. Inhibiting this enzyme is an efficient strategy to block viral replication. Structural studies provide valuable insight into the function of this protease and structural basis for rational inhibitor design. In this review, we describe structural studies on the main protease of SARS-CoV-2. The strategies applied in developing inhibitors of the main protease of SARS-CoV-2 and currently available protein inhibitors are summarized. Due to the availability of high-resolution structures, structure-guided drug design will play an important role in developing antivirals. The availability of high-resolution structures, potent peptidic inhibitors, and diverse compound scaffolds indicate the feasibility of developing potent protease inhibitors as antivirals for COVID-19.
Keywords: COVID-19; SARS-CoV-2; antivirals; drug discovery; protease inhibitor; protein structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
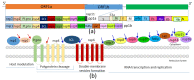

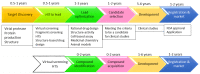

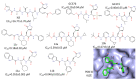
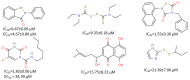
References
-
- Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T.D., Memish Z.A., Drosten C., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

