How Do Molecular Dynamics Data Complement Static Structural Data of GPCRs
- PMID: 32824756
- PMCID: PMC7460635
- DOI: 10.3390/ijms21165933
How Do Molecular Dynamics Data Complement Static Structural Data of GPCRs
Abstract
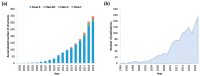
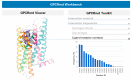
G protein-coupled receptors (GPCRs) are implicated in nearly every physiological process in the human body and therefore represent an important drug targeting class. Advances in X-ray crystallography and cryo-electron microscopy (cryo-EM) have provided multiple static structures of GPCRs in complex with various signaling partners. However, GPCR functionality is largely determined by their flexibility and ability to transition between distinct structural conformations. Due to this dynamic nature, a static snapshot does not fully explain the complexity of GPCR signal transduction. Molecular dynamics (MD) simulations offer the opportunity to simulate the structural motions of biological processes at atomic resolution. Thus, this technique can incorporate the missing information on protein flexibility into experimentally solved structures. Here, we review the contribution of MD simulations to complement static structural data and to improve our understanding of GPCR physiology and pharmacology, as well as the challenges that still need to be overcome to reach the full potential of this technique.
Keywords: GPCRs; drug discovery; ligand binding; molecular dynamics; receptor (in)activation; receptor signaling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

