The Quiet and Underappreciated Rise of Drug-Resistant Invasive Fungal Pathogens
- PMID: 32824785
- PMCID: PMC7557958
- DOI: 10.3390/jof6030138
The Quiet and Underappreciated Rise of Drug-Resistant Invasive Fungal Pathogens
Abstract
Human fungal pathogens are attributable to a significant economic burden and mortality worldwide. Antifungal treatments, although limited in number, play a pivotal role in decreasing mortality and morbidities posed by invasive fungal infections (IFIs). However, the recent emergence of multidrug-resistant Candida auris and Candida glabrata and acquiring invasive infections due to azole-resistant C. parapsilosis, C. tropicalis, and Aspergillus spp. in azole-naïve patients pose a serious health threat considering the limited number of systemic antifungals available to treat IFIs. Although advancing for major fungal pathogens, the understanding of fungal attributes contributing to antifungal resistance is just emerging for several clinically important MDR fungal pathogens. Further complicating the matter are the distinct differences in antifungal resistance mechanisms among various fungal species in which one or more mechanisms may contribute to the resistance phenotype. In this review, we attempt to summarize the burden of antifungal resistance for selected non-albicansCandida and clinically important Aspergillus species together with their phylogenetic placement on the tree of life. Moreover, we highlight the different molecular mechanisms between antifungal tolerance and resistance, and comprehensively discuss the molecular mechanisms of antifungal resistance in a species level.
Keywords: Aspergillus fumigatus; Aspergillus terreus; Candida auris; Candida glabrata; Candida parapsilosis; Candida tropicalis; antifungal resistance mechanisms.
Conflict of interest statement
D.S.P. receives research support and/or serves on advisory boards for Amplyx, Cidara, Scynexis, N8 Medical, Merck, Regeneron, and Pfizer. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


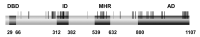

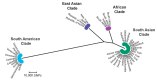
References
-
- Zhai B., Ola M., Rolling T., Tosini N.L., Joshowitz S., Littmann E.R., Amoretti L.A., Fontana E., Wright R.J., Miranda E., et al. High-resolution mycobiota analysis reveals dynamic intestinal translocation preceding invasive candidiasis. Nat. Med. 2020;26:59–64. doi: 10.1038/s41591-019-0709-7. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

