Distinct Mutation Patterns Reveal Melanoma Subtypes and Influence Immunotherapy Response in Advanced Melanoma Patients
- PMID: 32825510
- PMCID: PMC7563780
- DOI: 10.3390/cancers12092359
Distinct Mutation Patterns Reveal Melanoma Subtypes and Influence Immunotherapy Response in Advanced Melanoma Patients
Abstract
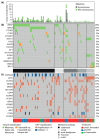
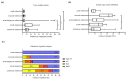
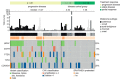
The detection of somatic driver mutations by next-generation sequencing (NGS) is becoming increasingly important in the care of advanced melanoma patients. In our study, we evaluated the NGS results of 82 melanoma patients from clinical routine in 2017. Besides determining the tumor mutational burden (TMB) and annotation of all genetic driver alterations, we investigated their potential as a predictor for resistance to immune checkpoint inhibitors (ICI) and as a distinguishing feature between melanoma subtypes. Melanomas of unknown primary had a similar mutation pattern and TMB to cutaneous melanoma, which hints at its cutaneous origin. Besides the typical hotspot mutation in BRAF and NRAS, we frequently observed CDKN2A deletions. Acral and mucosal melanomas were dominated by CNV alterations affecting PDGFRA, KIT, CDK4, RICTOR, CCND2 and CHEK2. Uveal melanoma often had somatic SNVs in GNA11/Q and amplification of MYC in all cases. A significantly higher incidence of BRAF V600 mutations and EGFR amplifications, PTEN and TP53 deletions was found in patients with disease progression while on ICI. Thus, NGS might help to characterize melanoma subtypes more precisely and to identify possible resistance mechanisms to ICI therapy. Nevertheless, NGS based studies, including larger cohorts, are needed to support potential genetic ICI resistance mechanisms.
Keywords: Genome of advanced melanoma; TMB; acral; immune checkpoint inhibitors; melanoma of unknown origin; mucosal; next-generation sequencing; tumor mutation burden; uveal.
Conflict of interest statement
A.F. served as consultant to Roche, Novartis, MSD, Pierre-Fabre; received travel support from Roche, Novartis, BMS, Pierre-Fabre, received speaker fees from Roche, Novartis, BMS, MSD and CeGaT, received research funding from BMS outside the submitted work. T.A. reports personal fees and travel grants from BMS, grants, personal fees and travel grants from Novartis, personal fees from Pierre Fabre and CeCaVa, grants from Neracare, grants from Sanofi, outside the submitted work. I.B. received speaker fees from Novartis and AstraZeneca and honoraria for advisory board participation from BMS and Novartis. CG reports grants and personal fees from Novartis, BMS, Roche, personal fees from MSD. Personal fees from Amgen, Philogen, LEO, Incyte, outside the submitted work. F.J.H., O.R. and C.S. received an institutional grant from Novartis. C.S. received research funding from BMS outside the submitted work. T.S. received grants from Novartis and Pierre-Fabre and personal fees from Neracare. The other authors declared no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

