Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin
- PMID: 32825611
- PMCID: PMC7551084
- DOI: 10.3390/toxins12090538
Primary Sequence and 3D Structure Prediction of the Plant Toxin Stenodactylin
Abstract
Stenodactylin is one of the most potent type 2 ribosome-inactivating proteins (RIPs); its high toxicity has been demonstrated in several models both in vitro and in vivo. Due to its peculiarities, stenodactylin could have several medical and biotechnological applications in neuroscience and cancer treatment. In this work, we report the complete amino acid sequence of stenodactylin and 3D structure prediction. The comparison between the primary sequence of stenodactylin and other RIPs allowed us to identify homologies/differences and the amino acids involved in RIP toxic activity. Stenodactylin RNA was isolated from plant caudex, reverse transcribed through PCR and the cDNA was amplificated and cloned into a plasmid vector and further analyzed by sequencing. Nucleotide sequence analysis showed that stenodactylin A and B chains contain 251 and 258 amino acids, respectively. The key amino acids of the active site described for ricin and most other RIPs are also conserved in the stenodactylin A chain. Stenodactylin amino acid sequence shows a high identity degree with volkensin (81.7% for A chain, 90.3% for B chain), whilst when compared with other type 2 RIPs the identity degree ranges from 27.7 to 33.0% for the A chain and from 42.1 to 47.7% for the B chain.
Keywords: 3D structure; plant toxin; primary sequence; ribosome-inactivating protein; stenodactylin; toxic lectin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




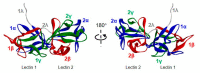



Similar articles
-
Sequence, Structure, and Binding Site Analysis of Kirkiin in Comparison with Ricin and Other Type 2 RIPs.Toxins (Basel). 2021 Dec 3;13(12):862. doi: 10.3390/toxins13120862. Toxins (Basel). 2021. PMID: 34941700 Free PMC article.
-
Binding and intracellular routing of the plant-toxic lectins, lanceolin and stenodactylin.Biochim Biophys Acta. 2010 Dec;1800(12):1276-82. doi: 10.1016/j.bbagen.2010.09.006. Epub 2010 Oct 7. Biochim Biophys Acta. 2010. PMID: 20933061
-
Crystallization and preliminary X-ray diffraction data analysis of stenodactylin, a highly toxic type 2 ribosome-inactivating protein from Adenia stenodactyla.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Jan 1;66(Pt 1):51-3. doi: 10.1107/S1744309109047654. Epub 2009 Dec 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20057070 Free PMC article.
-
Non-toxic type 2 ribosome-inactivating proteins (RIPs) from Sambucus: occurrence, cellular and molecular activities and potential uses.Cell Mol Biol (Noisy-le-grand). 2003 Jun;49(4):537-45. Cell Mol Biol (Noisy-le-grand). 2003. PMID: 12899446 Review.
-
Description, distribution, activity and phylogenetic relationship of ribosome-inactivating proteins in plants, fungi and bacteria.Mini Rev Med Chem. 2004 Jun;4(5):461-76. doi: 10.2174/1389557043403891. Mini Rev Med Chem. 2004. PMID: 15180503 Review.
Cited by
-
A Simple, Fast and Portable Method for Electrochemical Detection of Adenine Released by Ricin Enzymatic Activity.Toxins (Basel). 2021 Mar 26;13(4):238. doi: 10.3390/toxins13040238. Toxins (Basel). 2021. PMID: 33810228 Free PMC article.
-
Toxin and Immunotoxin Based Therapeutic Approaches.Toxins (Basel). 2022 Jan 17;14(1):63. doi: 10.3390/toxins14010063. Toxins (Basel). 2022. PMID: 35051040 Free PMC article.
-
Structure and Biological Properties of Ribosome-Inactivating Proteins and Lectins from Elder (Sambucus nigra L.) Leaves.Toxins (Basel). 2022 Sep 1;14(9):611. doi: 10.3390/toxins14090611. Toxins (Basel). 2022. PMID: 36136551 Free PMC article.
-
Sequence, Structure, and Binding Site Analysis of Kirkiin in Comparison with Ricin and Other Type 2 RIPs.Toxins (Basel). 2021 Dec 3;13(12):862. doi: 10.3390/toxins13120862. Toxins (Basel). 2021. PMID: 34941700 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

