Biochemical and functional characterization of a meiosis-specific Pch2/ORC AAA+ assembly
- PMID: 32826290
- PMCID: PMC7442955
- DOI: 10.26508/lsa.201900630
Biochemical and functional characterization of a meiosis-specific Pch2/ORC AAA+ assembly
Abstract
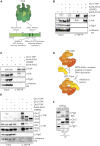
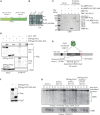
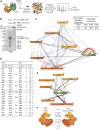
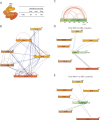
Pch2 is a meiosis-specific AAA+ protein that controls several important chromosomal processes. We previously demonstrated that Orc1, a subunit of the ORC, functionally interacts with budding yeast Pch2. The ORC (Orc1-6) AAA+ complex loads the AAA+ MCM helicase to origins of replication, but whether and how ORC collaborates with Pch2 remains unclear. Here, we show that a Pch2 hexamer directly associates with ORC during the meiotic G2/prophase. Biochemical analysis suggests that Pch2 uses its non-enzymatic NH2-terminal domain and AAA+ core and likely engages the interface of ORC that also binds to Cdc6, a factor crucial for ORC-MCM binding. Canonical ORC function requires association with origins, but we show here that despite causing efficient removal of Orc1 from origins, nuclear depletion of Orc2 and Orc5 does not trigger Pch2/Orc1-like meiotic phenotypes. This suggests that the function for Orc1/Pch2 in meiosis can be executed without efficient association of ORC with origins of replication. In conclusion, we uncover distinct functionalities for Orc1/ORC that drive the establishment of a non-canonical, meiosis-specific AAA+ assembly with Pch2.
© 2020 Villar-Fernández et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
