Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA4
- PMID: 32826317
- PMCID: PMC7606696
- DOI: 10.1074/jbc.RA120.014099
Csx3 is a cyclic oligonucleotide phosphodiesterase associated with type III CRISPR-Cas that degrades the second messenger cA4
Abstract
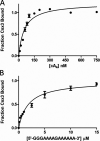
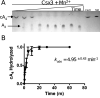
Cas10 is the signature gene for type III CRISPR-Cas surveillance complexes. Unlike type I and type II systems, type III systems do not require a protospacer adjacent motif and target nascent RNA associated with transcriptionally active DNA. Further, target RNA recognition activates the cyclase domain of Cas10, resulting in the synthesis of cyclic oligoadenylate second messengers. These second messengers are recognized by ancillary Cas proteins harboring CRISPR-associated Rossmann fold (CARF) domains and regulate the activities of these proteins in response to invading nucleic acid. Csx3 is a distant member of the CARF domain superfamily previously characterized as a Mn2+-dependent deadenylation exoribonuclease. However, its specific role in CRISPR-Cas defense remains to be determined. Here we show that Csx3 is strongly associated with type III systems and that Csx3 binds cyclic tetra-adenylate (cA4) second messenger with high affinity. Further, Csx3 harbors cyclic oligonucleotide phosphodiesterase activity that quickly degrades this cA4 signal. In addition, structural analysis identifies core elements that define the CARF domain fold, and the mechanistic basis for ring nuclease activity is discussed. Overall, the work suggests that Csx3 functions within CRISPR-Cas as a counterbalance to Cas10 to regulate the duration and amplitude of the cA4 signal, providing an off ramp from the programmed cell death pathway in cells that successfully cure viral infection.
Keywords: CARF; CRISPR/Cas; RNase; X-ray crystallography; cyclic nucleotide; cyclic tetra-adenylate; fluorescence; fluorescence lifetime; phosphodiesterase; phosphodiesterases.
© 2020 Brown et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures
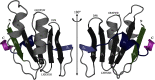
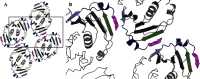

Similar articles
-
Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.Elife. 2020 Jun 29;9:e57627. doi: 10.7554/eLife.57627. Elife. 2020. PMID: 32597755 Free PMC article.
-
Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.Nucleic Acids Res. 2021 Dec 2;49(21):12577-12590. doi: 10.1093/nar/gkab1130. Nucleic Acids Res. 2021. PMID: 34850143 Free PMC article.
-
Structural basis of cyclic oligoadenylate binding to the transcription factor Csa3 outlines cross talk between type III and type I CRISPR systems.J Biol Chem. 2022 Feb;298(2):101591. doi: 10.1016/j.jbc.2022.101591. Epub 2022 Jan 14. J Biol Chem. 2022. PMID: 35038453 Free PMC article.
-
The diverse arsenal of type III CRISPR-Cas-associated CARF and SAVED effectors.Biochem Soc Trans. 2022 Oct 31;50(5):1353-1364. doi: 10.1042/BST20220289. Biochem Soc Trans. 2022. PMID: 36282000 Free PMC article. Review.
-
Type III CRISPR-Cas: beyond the Cas10 effector complex.Trends Biochem Sci. 2024 Jan;49(1):28-37. doi: 10.1016/j.tibs.2023.10.006. Epub 2023 Nov 8. Trends Biochem Sci. 2024. PMID: 37949766 Free PMC article. Review.
Cited by
-
Inhibitors of Cyclic Dinucleotide Phosphodiesterases and Cyclic Oligonucleotide Ring Nucleases as Potential Drugs for Various Diseases.Cells. 2025 Apr 30;14(9):663. doi: 10.3390/cells14090663. Cells. 2025. PMID: 40358186 Free PMC article. Review.
-
CRISPR Element Patterns vs. Pathoadaptability of Clinical Pseudomonas aeruginosa Isolates from a Medical Center in Moscow, Russia.Antibiotics (Basel). 2021 Oct 26;10(11):1301. doi: 10.3390/antibiotics10111301. Antibiotics (Basel). 2021. PMID: 34827239 Free PMC article.
-
Cyclic oligoadenylate signalling and regulation by ring nucleases during type III CRISPR defence.RNA. 2021 May 13;27(8):855-67. doi: 10.1261/rna.078739.121. Online ahead of print. RNA. 2021. PMID: 33986148 Free PMC article.
-
Genomic Distribution of Pro-Virulent cpdB-like Genes in Eubacteria and Comparison of the Enzyme Specificity of CpdB-like Proteins from Salmonella enterica, Escherichia coli and Streptococcus suis.Int J Mol Sci. 2023 Feb 19;24(4):4150. doi: 10.3390/ijms24044150. Int J Mol Sci. 2023. PMID: 36835561 Free PMC article.
-
Time-Dependent Fluorescence Spectroscopy to Quantify Complex Binding Interactions.ACS Omega. 2020 Nov 6;5(45):29017-29024. doi: 10.1021/acsomega.0c03416. eCollection 2020 Nov 17. ACS Omega. 2020. PMID: 33225133 Free PMC article.
References
-
- Lintner N. G., Frankel K. A., Tsutakawa S. E., Alsbury D. L., Copié V., Young M. J., Tainer J. A., and Lawrence C. M. (2011) The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J. Mol. Biol. 405, 939–955 10.1016/j.jmb.2010.11.019 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

