Hierarchical natural move Monte Carlo refines flexible RNA structures into cryo-EM densities
- PMID: 32826323
- PMCID: PMC7668250
- DOI: 10.1261/rna.071100.119
Hierarchical natural move Monte Carlo refines flexible RNA structures into cryo-EM densities
Abstract
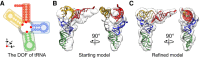
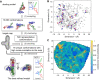
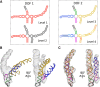
Ribonucleic acids (RNAs) play essential roles in living cells. Many of them fold into defined three-dimensional (3D) structures to perform functions. Recent advances in single-particle cryo-electron microscopy (cryo-EM) have enabled structure determinations of RNA to atomic resolutions. However, most RNA molecules are structurally flexible, limiting the resolution of their structures solved by cryo-EM. In modeling these molecules, several computational methods are limited by the requirement of massive computational resources and/or the low efficiency in exploring large-scale structural variations. Here we use hierarchical natural move Monte Carlo (HNMMC), which takes advantage of collective motions for groups of nucleic acid residues, to refine RNA structures into their cryo-EM maps, preserving atomic details in the models. After validating the method on a simulated density map of tRNA, we applied it to objectively obtain the model of the folding intermediate for the specificity domain of ribonuclease P from Bacillus subtilis and refine a flexible ribosomal RNA (rRNA) expansion segment from the Mycobacterium tuberculosis (Mtb) ribosome in different conformational states. Finally, we used HNMMC to model atomic details and flexibility for two distinct conformations of the complete genomic RNA (gRNA) inside MS2, a single-stranded RNA virus, revealing multiple pathways for its capsid assembly.
Keywords: RNA folding; RNA virus; conformational sampling; molecular motions; structural modeling.
© 2020 Chang et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

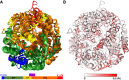
Similar articles
-
Structure of the E. coli ribosome-EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM.Nature. 2015 Apr 23;520(7548):567-70. doi: 10.1038/nature14275. Epub 2015 Feb 23. Nature. 2015. PMID: 25707802
-
Structure of the bacterial ribosome at 2 Å resolution.Elife. 2020 Sep 14;9:e60482. doi: 10.7554/eLife.60482. Elife. 2020. PMID: 32924932 Free PMC article.
-
Direct three-dimensional localization and positive identification of RNA helices within the ribosome by means of genetic tagging and cryo-electron microscopy.Structure. 1999 Dec 15;7(12):1567-73. doi: 10.1016/s0969-2126(00)88347-1. Structure. 1999. PMID: 10647187
-
Three-dimensional electron cryomicroscopy of ribosomes.Curr Protein Pept Sci. 2002 Feb;3(1):79-91. doi: 10.2174/1389203023380873. Curr Protein Pept Sci. 2002. PMID: 12370013 Review.
-
Almost lost in translation. Cryo-EM of a dynamic macromolecular complex: the ribosome.Eur Biophys J. 2011 May;40(5):589-97. doi: 10.1007/s00249-011-0683-6. Epub 2011 Feb 19. Eur Biophys J. 2011. PMID: 21336521 Review.
Cited by
-
Structural Assembly of Qβ Virion and Its Diverse Forms of Virus-like Particles.Viruses. 2022 Jan 24;14(2):225. doi: 10.3390/v14020225. Viruses. 2022. PMID: 35215818 Free PMC article.
-
Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages.Viruses. 2023 Sep 23;15(10):1985. doi: 10.3390/v15101985. Viruses. 2023. PMID: 37896763 Free PMC article. Review.
-
Transcriptional Riboswitches Integrate Timescales for Bacterial Gene Expression Control.Front Mol Biosci. 2021 Jan 13;7:607158. doi: 10.3389/fmolb.2020.607158. eCollection 2020. Front Mol Biosci. 2021. PMID: 33521053 Free PMC article. Review.
-
Removal of Pseudomonas type IV pili by a small RNA virus.Science. 2024 Apr 5;384(6691):eadl0635. doi: 10.1126/science.adl0635. Epub 2024 Apr 5. Science. 2024. PMID: 38574145 Free PMC article.
-
Structural basis of Acinetobacter type IV pili targeting by an RNA virus.Nat Commun. 2024 Mar 29;15(1):2746. doi: 10.1038/s41467-024-47119-5. Nat Commun. 2024. PMID: 38553443 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
