A cross-reactive human IgA monoclonal antibody blocks SARS-CoV-2 spike-ACE2 interaction
- PMID: 32826914
- PMCID: PMC7442812
- DOI: 10.1038/s41467-020-18058-8
A cross-reactive human IgA monoclonal antibody blocks SARS-CoV-2 spike-ACE2 interaction
Abstract
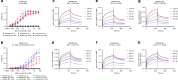
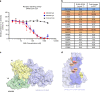
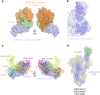
COVID-19 caused by SARS-CoV-2 has become a global pandemic requiring the development of interventions for the prevention or treatment to curtail mortality and morbidity. No vaccine to boost mucosal immunity, or as a therapeutic, has yet been developed to SARS-CoV-2. In this study, we discover and characterize a cross-reactive human IgA monoclonal antibody, MAb362. MAb362 binds to both SARS-CoV and SARS-CoV-2 spike proteins and competitively blocks ACE2 receptor binding, by overlapping the ACE2 structural binding epitope. Furthermore, MAb362 IgA neutralizes both pseudotyped SARS-CoV and SARS-CoV-2 in 293 cells expressing ACE2. When converted to secretory IgA, MAb326 also neutralizes authentic SARS-CoV-2 virus while the IgG isotype shows no neutralization. Our results suggest that SARS-CoV-2 specific IgA antibodies, such as MAb362, may provide effective immunity against SARS-CoV-2 by inducing mucosal immunity within the respiratory system, a potentially critical feature of an effective vaccine.
Conflict of interest statement
A patent application has been filed on 5 May 2020 on monoclonal antibodies targeting SARS-CoV-2 (U.S. Patent and Trademark Office patent application no. 63/020,483; patent applicants: Y.W., M.E., Q.L., and M.K., University of Massachusetts Medical School). The remaining authors declare no competing interests.
Figures

Update of
-
IgA MAb blocks SARS-CoV-2 Spike-ACE2 interaction providing mucosal immunity.bioRxiv [Preprint]. 2020 May 15:2020.05.15.096719. doi: 10.1101/2020.05.15.096719. bioRxiv. 2020. Update in: Nat Commun. 2020 Aug 21;11(1):4198. doi: 10.1038/s41467-020-18058-8. PMID: 32511396 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

