A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization
- PMID: 32827125
- PMCID: PMC7441844
- DOI: 10.1007/s15010-020-01503-7
A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization
Abstract
Objective: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic challenges national health systems and the global economy. Monitoring of infection rates and seroprevalence can guide public health measures to combat the pandemic. This depends on reliable tests on active and former infections. Here, we set out to develop and validate a specific and sensitive enzyme linked immunosorbent assay (ELISA) for detection of anti-SARS-CoV-2 antibody levels.
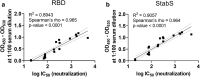
Methods: In our ELISA, we used SARS-CoV-2 receptor-binding domain (RBD) and a stabilized version of the spike (S) ectodomain as antigens. We assessed sera from patients infected with seasonal coronaviruses, SARS-CoV-2 and controls. We determined and monitored IgM-, IgA- and IgG-antibody responses towards these antigens. In addition, for a panel of 22 sera, virus neutralization and ELISA parameters were measured and correlated.
Results: The RBD-based ELISA detected SARS-CoV-2-directed antibodies, did not cross-react with seasonal coronavirus antibodies and correlated with virus neutralization (R2 = 0.89). Seroconversion started at 5 days after symptom onset and led to robust antibody levels at 10 days after symptom onset. We demonstrate high specificity (99.3%; N = 1000) and sensitivity (92% for IgA, 96% for IgG and 98% for IgM; > 10 days after PCR-proven infection; N = 53) in serum.
Conclusions: With the described RBD-based ELISA protocol, we provide a reliable test for seroepidemiological surveys. Due to high specificity and strong correlation with virus neutralization, the RBD ELISA holds great potential to become a preferred tool to assess thresholds of protective immunity after infection and vaccination.
Keywords: Antibody test; Assay validation; COVID-19; ELISA; Receptor binding domain; S protein; SARS-CoV-2; Serology; Spike protein; Virus neutralization.
Conflict of interest statement
All authors declare no conflicts of interest.
Figures




References
-
- van der Hoek L. Human coronaviruses: what do they cause? Antivir Ther. 2007;12:651–658. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

