Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation
- PMID: 32828819
- PMCID: PMC7725941
- DOI: 10.1053/j.gastro.2020.08.029
Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation
Erratum in
-
Correction.Gastroenterology. 2022 Jul;163(1):339. doi: 10.1053/j.gastro.2022.05.025. Epub 2022 May 25. Gastroenterology. 2022. PMID: 35643183 No abstract available.
Abstract
Background & aims: Countries endemic for parasitic infestations have a lower incidence of Crohn's disease (CD) than nonendemic countries, and there have been anecdotal reports of the beneficial effects of helminths in CD patients. Tuft cells in the small intestine sense and direct the immune response against eukaryotic parasites. We investigated the activities of tuft cells in patients with CD and mouse models of intestinal inflammation.
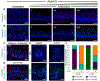
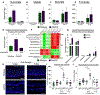
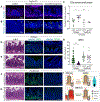
Methods: We used microscopy to quantify tuft cells in intestinal specimens from patients with ileal CD (n = 19), healthy individuals (n = 14), and TNFΔARE/+ mice, which develop Crohn's-like ileitis. We performed single-cell RNA sequencing, mass spectrometry, and microbiome profiling of intestinal tissues from wild-type and Atoh1-knockout mice, which have expansion of tuft cells, to study interactions between microbes and tuft cell populations. We assessed microbe dependence of tuft cell populations using microbiome depletion, organoids, and microbe transplant experiments. We used multiplex imaging and cytokine assays to assess alterations in inflammatory response following expansion of tuft cells with succinate administration in TNFΔARE/+ and anti-CD3E CD mouse models.
Results: Inflamed ileal tissues from patients and mice had reduced numbers of tuft cells, compared with healthy individuals or wild-type mice. Expansion of tuft cells was associated with increased expression of genes that regulate the tricarboxylic acid cycle, which resulted from microbe production of the metabolite succinate. Experiments in which we manipulated the intestinal microbiota of mice revealed the existence of an ATOH1-independent population of tuft cells that was sensitive to metabolites produced by microbes. Administration of succinate to mice expanded tuft cells and reduced intestinal inflammation in TNFΔARE/+ mice and anti-CD3E-treated mice, increased GATA3+ cells and type 2 cytokines (IL22, IL25, IL13), and decreased RORGT+ cells and type 17 cytokines (IL23) in a tuft cell-dependent manner.
Conclusions: We found that tuft cell expansion reduced chronic intestinal inflammation in mice. Strategies to expand tuft cells might be developed for treatment of CD.
Keywords: IBD; epithelium; heterogeneity; metabolism.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest Statement
J.R.W. is the founder of Resphera Biosciences. All other authors declare no conflict of financial interests.
Figures




Comment in
-
Could a Small Population of Epithelial Cells Get "Tuft" With Crohn's Disease?Gastroenterology. 2020 Dec;159(6):2025-2027. doi: 10.1053/j.gastro.2020.09.037. Epub 2020 Oct 1. Gastroenterology. 2020. PMID: 33011174 Free PMC article. No abstract available.
References
-
- Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54.e42. - PubMed
-
- de Silva P, Korzenik J. The Changing Epidemiology of Inflammatory Bowel Disease: Identifying New High-risk Populations. Clin Gastroenterol Heptaol. 2015; 4:690–2. - PubMed
-
- Su J, Chen T, Ji X-Y, et al. IL-25 Downregulates Th1/Th17 Immune Response in an IL-10–Dependent Manner in Inflammatory Bowel Disease. Inflamm Bowel Dis 2013;19:720–728. - PubMed
-
- Broadhurst MJ, Leung JM, Kashyap V, et al. IL-22+ CD4+ T cells are associated with therapeutic trichuris trichiura infection in an ulcerative colitis patient. Sci Transl Med 2010;2:60ra88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 LM012412/LM/NLM NIH HHS/United States
- P01 CA028842/CA/NCI NIH HHS/United States
- R01 DK128200/DK/NIDDK NIH HHS/United States
- K01 DK106311/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- P01 CA116087/CA/NCI NIH HHS/United States
- I01 BX001453/BX/BLRD VA/United States
- I01 CX002171/CX/CSRD VA/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- F31 GM120940/GM/NIGMS NIH HHS/United States
- R01 DK103831/DK/NIDDK NIH HHS/United States
- T32 HD007502/HD/NICHD NIH HHS/United States
- KL2 TR002245/TR/NCATS NIH HHS/United States
- R35 CA197570/CA/NCI NIH HHS/United States
- R21 AI142042/AI/NIAID NIH HHS/United States
- T32 AI138932/AI/NIAID NIH HHS/United States
- UM1 CA183727/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

