Brain connectivity alteration detection via matrix-variate differential network model
- PMID: 32829503
- PMCID: PMC7900256
- DOI: 10.1111/biom.13359
Brain connectivity alteration detection via matrix-variate differential network model
Abstract
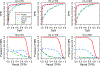
Brain functional connectivity reveals the synchronization of brain systems through correlations in neurophysiological measures of brain activities. Growing evidence now suggests that the brain connectivity network experiences alterations with the presence of numerous neurological disorders, thus differential brain network analysis may provide new insights into disease pathologies. The data from neurophysiological measurement are often multidimensional and in a matrix form, posing a challenge in brain connectivity analysis. Existing graphical model estimation methods either assume a vector normal distribution that in essence requires the columns of the matrix data to be independent or fail to address the estimation of differential networks across different populations. To tackle these issues, we propose an innovative matrix-variate differential network (MVDN) model. We exploit the D-trace loss function and a Lasso-type penalty to directly estimate the spatial differential partial correlation matrix and use an alternating direction method of multipliers algorithm for the optimization problem. Theoretical and simulation studies demonstrate that MVDN significantly outperforms other state-of-the-art methods in dynamic differential network analysis. We illustrate with a functional connectivity analysis of an attention deficit hyperactivity disorder dataset. The hub nodes and differential interaction patterns identified are consistent with existing experimental studies.
Keywords: brain network; differential network analysis; fMRI; graphical model; matrix data; variable selection.
© 2020 The International Biometric Society.
Figures
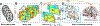
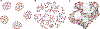



Similar articles
-
Simultaneous cluster structure learning and estimation of heterogeneous graphs for matrix-variate fMRI data.Biometrics. 2023 Sep;79(3):2246-2259. doi: 10.1111/biom.13753. Epub 2022 Sep 13. Biometrics. 2023. PMID: 36017603 Free PMC article.
-
Simultaneous differential network analysis and classification for matrix-variate data with application to brain connectivity.Biostatistics. 2022 Jul 18;23(3):967-989. doi: 10.1093/biostatistics/kxab007. Biostatistics. 2022. PMID: 33769450 Free PMC article.
-
Ordinal Pattern: A New Descriptor for Brain Connectivity Networks.IEEE Trans Med Imaging. 2018 Jul;37(7):1711-1722. doi: 10.1109/TMI.2018.2798500. IEEE Trans Med Imaging. 2018. PMID: 29969421
-
A survey on applications and analysis methods of functional magnetic resonance imaging for Alzheimer's disease.J Neurosci Methods. 2019 Apr 1;317:121-140. doi: 10.1016/j.jneumeth.2018.12.012. Epub 2018 Dec 26. J Neurosci Methods. 2019. PMID: 30593787 Review.
-
Different functional alteration in attention-deficit/hyperactivity disorder across developmental age groups: A meta-analysis and an independent validation of resting-state functional connectivity studies.CNS Neurosci Ther. 2023 Jan;29(1):60-69. doi: 10.1111/cns.14032. Epub 2022 Dec 5. CNS Neurosci Ther. 2023. PMID: 36468409 Free PMC article. Review.
Cited by
-
Simultaneous cluster structure learning and estimation of heterogeneous graphs for matrix-variate fMRI data.Biometrics. 2023 Sep;79(3):2246-2259. doi: 10.1111/biom.13753. Epub 2022 Sep 13. Biometrics. 2023. PMID: 36017603 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

