Developmental stage-associated microbiota profile of the peach fruit fly, Bactrocera zonata (Diptera: Tephritidae) and their functional prediction using 16S rRNA gene metabarcoding sequencing
- PMID: 32832340
- PMCID: PMC7426347
- DOI: 10.1007/s13205-020-02381-4
Developmental stage-associated microbiota profile of the peach fruit fly, Bactrocera zonata (Diptera: Tephritidae) and their functional prediction using 16S rRNA gene metabarcoding sequencing
Abstract
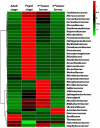
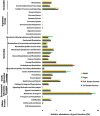
The different developmental stage-associated microbiota of the peach fruit fly, Bactrocera zonata (Diptera: Tephritidae), was characterized using 16S rRNA gene (V3-V4 region) metabarcoding on the Illumina HiSeq platform. Taxonomically, at 97% similarity, there were total 16 bacterial phyla, comprising of 24 classes, 55 orders, 90 families and 134 genera. Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes were the most abundant phyla with Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, Bacteroidia and Bacilli being the most abundant classes. The bacterial genus Enterobacter was dominant in the larval and adult stages and Pseudomonas in the pupal stage. A total of 2645 operational taxonomic units (OTUs) were identified, out of which 151 OTUs (core microbiota) were common among all the developmental stages of B. zonata. The genus Enterobacter, Klebsiella and Pantoea were dominant among the core microbiota. PICURSt analysis predicted that microbiota associated with B. zonata may be involved in membrane transport, carbohydrate metabolism, amino acid metabolism, replication and repair processes as well as in cellular processes and signalling. The microbiota that was shared by all the developmental stages of B. zonata in the present study could be targeted and the foundation for research on microbiota-based management of fruit flies.
Keywords: 16S rRNA gene; Bactrocera zonata; Management; Microbiota; Ontogeny.
© King Abdulaziz City for Science and Technology 2020.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures



References
-
- Abdelfattah A, Malacrinò A, Wisniewski M, Cacciola SO, Schena L. Metabarcoding: a powerful tool to investigate microbial communities and shape future plant protection strategies. Biol Control. 2017;120:1–10. doi: 10.1016/j.biocontrol.2017.07.009. - DOI
-
- Arasu MV, Duraipandiyan V, Agastian P, Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J Med Mycol. 2008;18:147–153.
LinkOut - more resources
Full Text Sources

