Dynamic allosteric communication pathway directing differential activation of the glucocorticoid receptor
- PMID: 32832645
- PMCID: PMC7439413
- DOI: 10.1126/sciadv.abb5277
Dynamic allosteric communication pathway directing differential activation of the glucocorticoid receptor
Abstract
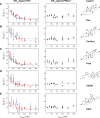
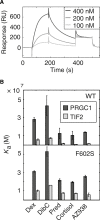
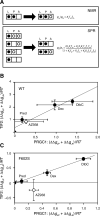
Allosteric communication within proteins is a hallmark of biochemical signaling, but the dynamic transmission pathways remain poorly characterized. We combined NMR spectroscopy and surface plasmon resonance to reveal these pathways and quantify their energetics in the glucocorticoid receptor, a transcriptional regulator controlling development, metabolism, and immune response. Our results delineate a dynamic communication network of residues linking the ligand-binding pocket to the activation function-2 interface, where helix 12, a switch for transcriptional activation, exhibits ligand- and coregulator-dependent dynamics coupled to graded activation. The allosteric free energy responds to variations in ligand structure: subtle changes gradually tune allostery while preserving the transmission pathway, whereas substitution of the entire pharmacophore leads to divergent allosteric control by apparently rewiring the communication network. Our results provide key insights that should aid in the design of mechanistically differentiated ligands.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Keenan C. R., Lew M. J., Stewart A. G., Biased signalling from the glucocorticoid receptor: Renewed opportunity for tailoring glucocorticoid activity. Biochem. Pharmacol. 112, 6–12 (2016). - PubMed
-
- Buttgereit F., Can we shift the benefit–risk ratio of glucocorticoids? Lancet Rheumatol. 2, E5–E6 (2020). - PubMed
-
- Bledsoe R. K., Montana V. G., Stanley T. B., Delves C. J., Apolito C. J., McKee D. D., Consler T. G., Parks D. J., Stewart E. L., Willson T. M., Lambert M. H., Moore J. T., Pearce K. H., Xu H. E., Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110, 93–105 (2002). - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

