Modeling and forecasting the COVID-19 pandemic in India
- PMID: 32834603
- PMCID: PMC7321056
- DOI: 10.1016/j.chaos.2020.110049
Modeling and forecasting the COVID-19 pandemic in India
Abstract
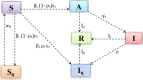
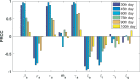
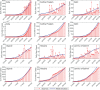
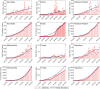
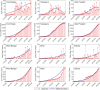
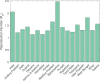
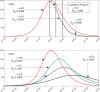
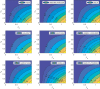
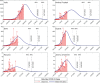
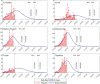
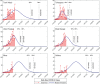
In India, 100,340 confirmed cases and 3155 confirmed deaths due to COVID-19 were reported as of May 18, 2020. Due to absence of specific vaccine or therapy, non-pharmacological interventions including social distancing, contact tracing are essential to end the worldwide COVID-19. We propose a mathematical model that predicts the dynamics of COVID-19 in 17 provinces of India and the overall India. A complete scenario is given to demonstrate the estimated pandemic life cycle along with the real data or history to date, which in turn divulges the predicted inflection point and ending phase of SARS-CoV-2. The proposed model monitors the dynamics of six compartments, namely susceptible (S), asymptomatic (A), recovered (R), infected (I), isolated infected (Iq ) and quarantined susceptible (Sq ), collectively expressed SARIIqSq . A sensitivity analysis is conducted to determine the robustness of model predictions to parameter values and the sensitive parameters are estimated from the real data on the COVID-19 pandemic in India. Our results reveal that achieving a reduction in the contact rate between uninfected and infected individuals by quarantined the susceptible individuals, can effectively reduce the basic reproduction number. Our model simulations demonstrate that the elimination of ongoing SARS-CoV-2 pandemic is possible by combining the restrictive social distancing and contact tracing. Our predictions are based on real data with reasonable assumptions, whereas the accurate course of epidemic heavily depends on how and when quarantine, isolation and precautionary measures are enforced.
Keywords: Basic reproduction number; COVID-19; Isolation; Mathematical model; Model prediction; Sensitivity analysis.
© 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- World Health Organization (WHO), Coronavirus disease (COVID-19) outbreak situation. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
-
- India COVID-19 tracker. 2020a. https://www.covid19india.org/.
-
- WHO Situation Report, Coronavirus disease 2019 (COVID-19). 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/2....
-
- BBC News. https://www.bbc.com/news/world-52114829.
LinkOut - more resources
Full Text Sources
Miscellaneous

