Intra-host Ebola viral adaption during human infection
- PMID: 32835207
- PMCID: PMC7347341
- DOI: 10.1016/j.bsheal.2019.02.001
Intra-host Ebola viral adaption during human infection
Abstract
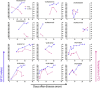
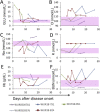
The onsite next generation sequencing (NGS) of Ebola virus (EBOV) genomes during the 2013-2016 Ebola epidemic in Western Africa provides an opportunity to trace the origin, transmission, and evolution of this virus. Herein, we have diagnosed a cohort of EBOV patients in Sierra Leone in 2015, during the late phase of the outbreak. The surviving EBOV patients had a recovery process characterized by decreasing viremia, fever, and biochemical parameters. EBOV genomes sequenced through the longitudinal blood samples of these patients showed dynamic intra-host substitutions of the virus during acute infection, including the previously described short stretches of 13 serial T>C mutations. Remarkably, within individual patients, samples collected during the early phase of infection possessed Ts at these nucleotide sites, whereas they were replaced by Cs in samples collected in the later phase, suggesting that these short stretches of T>C mutations could emerge independently. In addition, up to a total of 35 nucleotide sites spanning the EBOV genome were mutated coincidently. Our study showed the dynamic intra-host adaptation of EBOV during patient recovery and gave more insight into the complex EBOV-host interactions.
Keywords: Adaptation; Blood biochemistry; Clinical manifestations; Ebola virus; Genome sequencing; Intra-host Nucleotide Variation.
© 2019 Chinese Medical Association Publishing House. Published by Elsevier B.V.
Figures



References
-
- Tong Y.G., Shi W.F., Liu D., Qian J., Liang L., Bo X.C., Liu J., Ren H.G., Fan H., Ni M., Sun Y., Jin Y., Teng Y., Li Z., Kargbo D., Dafae F., Kanu A., Chen C.C., Lan Z.H., Jiang H., Luo Y., Lu H.J., Zhang X.G., Yang F., Hu Y., Cao Y.X., Deng Y.Q., Su H.X., Sun Y., Liu W.S., Wang Z., Wang C.Y., Bu Z.Y., Guo Z.D., Zhang L.B., Nie W.M., Bai C.Q., Sun C.H., An X.P., Xu P.S., Zhang X.L., Huang Y., Mi Z.Q., Yu D., Yao H.W., Feng Y., Xia Z.P., Zheng X.X., Yang S.T., Lu B., et al. Genetic diversity and evolutionary dynamics of Ebola virus in Sierra Leone. Nature. 2015;524:93–96. - PMC - PubMed
-
- Quick J., Loman N.J., Duraffour S., Simpson J.T., Severi E., Cowley L., Bore J.A., Koundouno R., Dudas G., Mikhail A., Ouedraogo N., Afrough B., Bah A., Baum J.H., Becker-Ziaja B., Boettcher J.P., Cabeza-Cabrerizo M., Camino-Sanchez A., Carter L.L., Doerrbecker J., Enkirch T., Garcia-Dorival I., Hetzelt N., Hinzmann J., Holm T., Kafetzopoulou L.E., Koropogui M., Kosgey A., Kuisma E., Logue C.H., Mazzarelli A., Meisel S., Mertens M., Michel J., Ngabo D., Nitzsche K., Pallasch E., Patrono L.V., Portmann J., Repits J.G., Rickett N.Y., Sachse A., Singethan K., Vitoriano I., Yemanaberhan R.L., Zekeng E.G., Racine T., Bello A., Sall A.A., Faye O., et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. - PMC - PubMed
-
- Ladner J.T., Wiley M.R., Mate S., Dudas G., Prieto K., Lovett S., Nagle E.R., Beitzel B., Gilbert M.L., Fakoli L., Diclaro J.W., II, Schoepp R.J., Fair J., Kuhn J.H., Hensley L.E., Park D.J., Sabeti P.C., Rambaut A., Sanchez-Lockhart M., Bolay F.K., Kugelman J.R., Palacios G. Evolution and spread of Ebola virus in Liberia, 2014–2015. Cell Host Microbe. 2015;18:659–669. - PMC - PubMed
-
- Park D.J., Dudas G., Wohl S., Goba A., Whitmer S.L., Andersen K.G., Sealfon R.S., Ladner J.T., Kugelman J.R., Matranga C.B., Winnicki S.M., Qu J., Gire S.K., Gladden-Young A., Jalloh S., Nosamiefan D., Yozwiak N.L., Moses L.M., Jiang P.P., Lin A.E., Schaffner S.F., Bird B., Towner J., Mamoh M., Gbakie M., Kanneh L., Kargbo D., Massally J.L., Kamara F.K., Konuwa E., Sellu J., Jalloh A.A., Mustapha I., Foday M., Yillah M., Erickson B.R., Sealy T., Blau D., Paddock C., Brault A., Amman B., Basile J., Bearden S., Belser J., Bergeron E., Campbell S., Chakrabarti A., Dodd K., Flint M., Gibbons A., et al. Ebola virus epidemiology, transmission, and evolution during seven months in Sierra Leone. Cell. 2015;161:1516–1526. - PMC - PubMed
-
- Gire S.K., Goba A., Andersen K.G., Sealfon R.S., Park D.J., Kanneh L., Jalloh S., Momoh M., Fullah M., Dudas G., Wohl S., Moses L.M., Yozwiak N.L., Winnicki S., Matranga C.B., Malboeuf C.M., Qu J., Gladden A.D., Schaffner S.F., Yang X., Jiang P.P., Nekoui M., Colubri A., Coomber M.R., Fonnie M., Moigboi A., Gbakie M., Kamara F.K., Tucker V., Konuwa E., Saffa S., Sellu J., Jalloh A.A., Kovoma A., Koninga J., Mustapha I., Kargbo K., Foday M., Yillah M., Kanneh F., Robert W., Massally J.L., Chapman S.B., Bochicchio J., Murphy C., Nusbaum C., Young S., Birren B.W., Grant D.S., Scheiffelin J.S., et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science. 2014;345:1369–1372. - PMC - PubMed
LinkOut - more resources
Full Text Sources

