Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health
- PMID: 32835590
- PMCID: PMC7524325
- DOI: 10.1080/19490976.2020.1802866
Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health
Abstract
Over the last two decades our understanding of the gut microbiota and its contribution to health and disease has been transformed. Among a new 'generation' of potentially beneficial microbes to have been recognized are members of the genus Eubacterium, who form a part of the core human gut microbiome. The genus consists of phylogenetically, and quite frequently phenotypically, diverse species, making Eubacterium a taxonomically unique and challenging genus. Several members of the genus produce butyrate, which plays a critical role in energy homeostasis, colonic motility, immunomodulation and suppression of inflammation in the gut. Eubacterium spp. also carry out bile acid and cholesterol transformations in the gut, thereby contributing to their homeostasis. Gut dysbiosis and a consequently modified representation of Eubacterium spp. in the gut, have been linked with various human disease states. This review provides an overview of Eubacterium species from a phylogenetic perspective, describes how they alter with diet and age and summarizes its association with the human gut and various health conditions.
Keywords: Eubacterium; Eubacterium hallii; Eubacterium rectale; bile acids; butyrate; cholesterol; gut microbiota; irritable bowel syndrome; phylogeny; short-chain fatty acids.
Figures

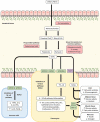
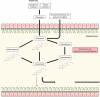

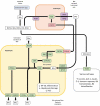
References
-
- Wade WG. The genus Eubacterium and related genera. Prokaryotes. 2006;4:823–835.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
