Recent insights for the emerging COVID-19: Drug discovery, therapeutic options and vaccine development
- PMID: 32837565
- PMCID: PMC7335243
- DOI: 10.1016/j.ajps.2020.06.001
Recent insights for the emerging COVID-19: Drug discovery, therapeutic options and vaccine development
Abstract
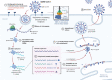
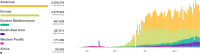
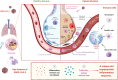
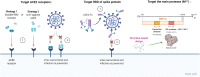
SARS-CoV-2 has been marked as a highly pathogenic coronavirus of COVID-19 disease into the human population, causing over 5.5 million confirmed cases worldwide. As COVID-19 has posed a global threat with significant human casualties and severe economic losses, there is a pressing demand to further understand the current situation and develop rational strategies to contain the drastic spread of the virus. Although there are no specific antiviral therapies that have proven effective in randomized clinical trials, currently, the rapid detection technology along with several promising therapeutics for COVID-19 have mitigated its drastic transmission. Besides, global institutions and corporations have commenced to parse out effective vaccines for the prevention of COVID-19. Herein, the present review will give exhaustive details of extensive researches concerning the drug discovery and therapeutic options for COVID-19 as well as some insightful discussions of the status of COVID-19.
Keywords: COVID-19; Coronavirus; Drug discovery; Precise prevention and control; SARS-CoV-2; Vaccine development.
© 2020 Shenyang Pharmaceutical University. Published by Elsevier B.V.
Conflict of interest statement
The authors assert that there is no conflict of interests concerning the publication of this review.
Figures




References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

