Molecular diagnostic technologies for COVID-19: Limitations and challenges
- PMID: 32837738
- PMCID: PMC7406419
- DOI: 10.1016/j.jare.2020.08.002
Molecular diagnostic technologies for COVID-19: Limitations and challenges
Abstract
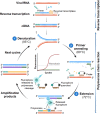
Background: To curb the spread of the COVID-19 (coronavirus disease 2019) pandemic, the world needs diagnostic systems capable of rapid detection and quantification of the novel coronavirus (SARS-CoV-2). Many biomedical companies are rising to the challenge and developing COVID-19 diagnostics. In the last few months, some of these diagnostics have become commercially available for healthcare workers and clinical laboratories. However, the diagnostic technologies have specific limitations and reported several false-positive and false-negative cases, especially during the early stages of infection.
Aim: This article aims to review recent developments in the field of COVID-19 diagnostics based on molecular technologies and analyze their clinical performance data.
Key concepts: The literature survey and performance-based analysis of the commercial and pre-commercial molecular diagnostics address several questions and issues related to the limitations of current technologies and highlight future research and development challenges to enable timely, rapid, low-cost, and accurate diagnosis of emerging infectious diseases.
Keywords: COVID-19; Clinical sensitivity; In vitro diagnostics; Point-of-care; Real-time RT-PCR; SARS-CoV-2.
© 2020 The Author. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The author declares that he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Baize S., Pannetier D., Oestereich L., Rieger T., Koivogui L., Magassouba N. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med. 2014;371:1418–1425. - PubMed
-
- World Health Organization W. Coronavirus disease 2019 (COVID-19): situation report, 72; 2020.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

