Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2
- PMID: 32837985
- PMCID: PMC7366953
- DOI: 10.1016/j.gendis.2020.07.006
Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2
Abstract
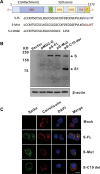
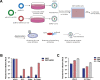
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative virus of the coronavirus disease 2019 (COVID-19) pandemic. To establish a safe and convenient assay system for studying entry inhibitors and neutralizing antibodies against SARS-CoV-2, we constructed a codon-optimized, full-length C-terminal mutant spike (S) gene of SARS-CoV-2. We generated a luciferase (Luc)-expressing pseudovirus containing the wild-type or mutant S protein of SARS-CoV-2 in the envelope-defective HIV-1 backbone. The key parameters for this pseudovirus-based assay, including the S mutants and virus incubation time, were optimized. This pseudovirus contains a Luc reporter gene that enabled us to easily quantify virus entry into angiotensin-converting enzyme 2 (ACE2)-expressing 293T cells. Cathepsin (Cat)B/L inhibitor E-64d could significantly block SARS-CoV-2 pseudovirus infection in 293T-ACE2 cells. Furthermore, the SARS-CoV-2 spike pseudotyped virus could be neutralized by sera from convalescent COVID-19 patients or recombinant ACE2 with the fused Fc region of human IgG1. Thus, we developed a pseudovirus-based assay for SARS-CoV-2, which will be valuable for evaluating viral entry inhibitors and neutralizing antibodies against this highly pathogenic virus.
Keywords: Antiviral therapeutics; Coronavirus; Neutralizing antibodies; Pseudovirus; SARS-CoV-2; Spike protein.
© 2020 Chongqing Medical University. Production and hosting by Elsevier B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

