SARS-CoV-2-human protein-protein interaction network
- PMID: 32838020
- PMCID: PMC7425553
- DOI: 10.1016/j.imu.2020.100413
SARS-CoV-2-human protein-protein interaction network
Abstract
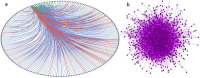
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the novel coronavirus which caused the coronavirus disease 2019 pandemic and infected more than 12 million victims and resulted in over 560,000 deaths in 213 countries around the world. Having no symptoms in the first week of infection increases the rate of spreading the virus. The increasing rate of the number of infected individuals and its high mortality necessitates an immediate development of proper diagnostic methods and effective treatments. SARS-CoV-2, similar to other viruses, needs to interact with the host proteins to reach the host cells and replicate its genome. Consequently, virus-host protein-protein interaction (PPI) identification could be useful in predicting the behavior of the virus and the design of antiviral drugs. Identification of virus-host PPIs using experimental approaches are very time consuming and expensive. Computational approaches could be acceptable alternatives for many preliminary investigations. In this study, we developed a new method to predict SARS-CoV-2-human PPIs. Our model is a three-layer network in which the first layer contains the most similar Alphainfluenzavirus proteins to SARS-CoV-2 proteins. The second layer contains protein-protein interactions between Alphainfluenzavirus proteins and human proteins. The last layer reveals protein-protein interactions between SARS-CoV-2 proteins and human proteins by using the clustering coefficient network property on the first two layers. To further analyze the results of our prediction network, we investigated human proteins targeted by SARS-CoV-2 proteins and reported the most central human proteins in human PPI network. Moreover, differentially expressed genes of previous researches were investigated and PPIs of SARS-CoV-2-human network, the human proteins of which were related to upregulated genes, were reported.
Keywords: COVID-19; Coronavirus; Host-pathogen protein interaction; Protein interaction prediction; Protein-protein interaction; SARS-CoV-2.
© 2020 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Khani H., Tabarraei A., Moradi A. Survey of coronaviruses infection among patients with flu-like symptoms in the golestan province, Iran TT - mljgoums. Nov. 2018;12(6):1–4.
-
- Qu J., Wickramasinghe C. SARS, MERS and the sunspot cycle. Curr Sci. 2017;113(8):1501.
-
- Organization W.H. 2003. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003.
LinkOut - more resources
Full Text Sources
Miscellaneous
