Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis
- PMID: 32838362
- PMCID: PMC7373048
- DOI: 10.1016/j.medj.2020.07.002
Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis
Abstract
Background: The ongoing coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a global public health concern due to relatively easy person-to-person transmission and the current lack of effective antiviral therapy. However, the exact molecular mechanisms of SARS-CoV-2 pathogenesis remain largely unknown.
Methods: Genome-wide screening was used to establish intraviral and viral-host interactomes. Quantitative proteomics was used to investigate the peripheral blood mononuclear cell (PBMC) proteome signature in COVID-19.
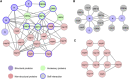
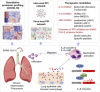
Findings: We elucidated 286 host proteins targeted by SARS-CoV-2 and >350 host proteins that are significantly perturbed in COVID-19-derived PBMCs. This signature in severe COVID-19 PBMCs reveals a significant upregulation of cellular proteins related to neutrophil activation and blood coagulation, as well as a downregulation of proteins mediating T cell receptor signaling. From the interactome, we further identified that non-structural protein 10 interacts with NF-κB-repressing factor (NKRF) to facilitate interleukin-8 (IL-8) induction, which potentially contributes to IL-8-mediated chemotaxis of neutrophils and the overexuberant host inflammatory response observed in COVID-19 patients.
Conclusions: Our study not only presents a systematic examination of SARS-CoV-2-induced perturbation of host targets and cellular networks but it also reveals insights into the mechanisms by which SARS-CoV-2 triggers cytokine storms, representing a powerful resource in the pursuit of therapeutic interventions.
Funding: National Key Research and Development Project of China, National Natural Science Foundation of China, National Science and Technology Major Project, Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, Shanghai Science and Technology Commission, Shanghai Municipal Health Commission, Shanghai Municipal Key Clinical Specialty, Innovative Research Team of High-level Local Universities in Shanghai, Interdisciplinary Program of Shanghai Jiao Tong University, SII Challenge Fund for COVID-19 Research, Chinese Academy of Sciences (CAS) Large Research Infrastructure of Maintenance and Remolding Project, and Chinese Academy of Sciences Key Technology Talent Program.
Keywords: COVID-19; IL-8; NKRF; SARS-CoV-2; interactome; nsp10; proteomics.
© 2020 Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

