Growth, detection, quantification, and inactivation of SARS-CoV-2
- PMID: 32838945
- PMCID: PMC7293183
- DOI: 10.1016/j.virol.2020.05.015
Growth, detection, quantification, and inactivation of SARS-CoV-2
Abstract
Severe acute respiratory syndrome coronavirus (SARS-CoV)-2 is the agent responsible for the coronavirus disease 2019 (COVID-19) global pandemic. SARS-CoV-2 is closely related to SARS-CoV, which caused the 2003 SARS outbreak. Although numerous reagents were developed to study SARS-CoV infections, few have been applicable to evaluating SARS-CoV-2 infection and immunity. Current limitations in studying SARS-CoV-2 include few validated assays with fully replication-competent wild-type virus. We have developed protocols to propagate, quantify, and work with infectious SARS-CoV-2. Here, we describe: (1) virus stock generation, (2) RT-qPCR quantification of SARS-CoV-2 RNA; (3) detection of SARS-CoV-2 antigen by flow cytometry, (4) quantification of infectious SARS-CoV-2 by focus-forming and plaque assays; and (5) validated protocols for virus inactivation. Collectively, these methods can be adapted to a variety of experimental designs, which should accelerate our understanding of SARS-CoV-2 biology and the development of effective countermeasures against COVID-19.
Keywords: Coronavirus; Flow cytometry; Focus-forming assay; Plaque assay; SARS-CoV-2; Titration; Virus inactivation.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and on the Scientific Advisory Board of Moderna.
Figures

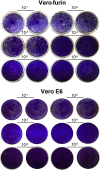

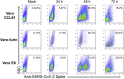
References
-
- (CDC), C.f.D.C.a.P . 2020. 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes.
-
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 - PMC - PubMed
-
- Joyce M.G., Sankhala R.S., Chen W.-H., Choe M., Bai H., Hajduczki A., Yan L., Sterling S.L., Peterson C.E., Green E.C., Smith C., de Val N., Amare M., Scott P., Laing E.D., Broder C.C., Rolland M., Michael N.L., Modjarrad K. 2020. A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. bioRxiv.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

