CAMSAP1 breaks the homeostatic microtubule network to instruct neuronal polarity
- PMID: 32839317
- PMCID: PMC7486724
- DOI: 10.1073/pnas.1913177117
CAMSAP1 breaks the homeostatic microtubule network to instruct neuronal polarity
Abstract
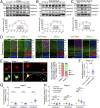
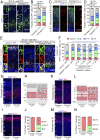
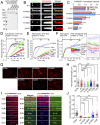
The establishment of axon/dendrite polarity is fundamental for neurons to integrate into functional circuits, and this process is critically dependent on microtubules (MTs). In the early stages of the establishment process, MTs in axons change dramatically with the morphological building of neurons; however, how the MT network changes are triggered is unclear. Here we show that CAMSAP1 plays a decisive role in the neuronal axon identification process by regulating the number of MTs. Neurons lacking CAMSAP1 form a multiple axon phenotype in vitro, while the multipolar-bipolar transition and radial migration are blocked in vivo. We demonstrate that the polarity regulator MARK2 kinase phosphorylates CAMSAP1 and affects its ability to bind to MTs, which in turn changes the protection of MT minus-ends and also triggers asymmetric distribution of MTs. Our results indicate that the polarized MT network in neurons is a decisive factor in establishing axon/dendritic polarity and is initially triggered by polarized signals.
Keywords: CAMSAP1; MARK2; cell migration; neuronal polarity; noncentrosomal microtubules.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Araújo S. J., Tear G., Axon guidance mechanisms and molecules: Lessons from invertebrates. Nat. Rev. Neurosci. 4, 910–922 (2003). - PubMed
-
- Shelly M., Poo M. M., Role of LKB1-SAD/MARK pathway in neuronal polarization. Dev. Neurobiol. 71, 508–527 (2011). - PubMed
-
- Kapitein L. C., Hoogenraad C. C., Building the neuronal microtubule cytoskeleton. Neuron 87, 492–506 (2015). - PubMed
-
- Conde C., Cáceres A., Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 10, 319–332 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

