Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae
- PMID: 32839345
- PMCID: PMC7502704
- DOI: 10.1073/pnas.2010214117
Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae
Abstract
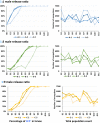
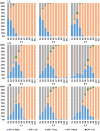
A Cas9/guide RNA-based gene drive strain, AgNosCd-1, was developed to deliver antiparasite effector molecules to the malaria vector mosquito, Anopheles gambiae The drive system targets the cardinal gene ortholog producing a red-eye phenotype. Drive can achieve 98 to 100% in both sexes and full introduction was observed in small cage trials within 6 to 10 generations following a single release of gene-drive males. No genetic load resulting from the integrated transgenes impaired drive performance in the trials. Potential drive-resistant target-site alleles arise at a frequency <0.1, and five of the most prevalent polymorphisms in the guide RNA target site in collections of colonized and wild-derived African mosquitoes do not prevent cleavage in vitro by the Cas9/guide RNA complex. Only one predicted off-target site is cleavable in vitro, with negligible deletions observed in vivo. AgNosCd-1 meets key performance criteria of a target product profile and can be a valuable component of a field-ready strain for mosquito population modification to control malaria transmission.
Keywords: cage trials; guide RNA polymorphisms; load; nontarget; off-target.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures