Spatial organization of Clostridium difficile S-layer biogenesis
- PMID: 32839524
- PMCID: PMC7445750
- DOI: 10.1038/s41598-020-71059-x
Spatial organization of Clostridium difficile S-layer biogenesis
Abstract
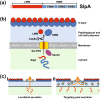
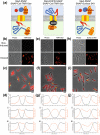
Surface layers (S-layers) are protective protein coats which form around all archaea and most bacterial cells. Clostridium difficile is a Gram-positive bacterium with an S-layer covering its peptidoglycan cell wall. The S-layer in C. difficile is constructed mainly of S-layer protein A (SlpA), which is a key virulence factor and an absolute requirement for disease. S-layer biogenesis is a complex multi-step process, disruption of which has severe consequences for the bacterium. We examined the subcellular localization of SlpA secretion and S-layer growth; observing formation of S-layer at specific sites that coincide with cell wall synthesis, while the secretion of SlpA from the cell is relatively delocalized. We conclude that this delocalized secretion of SlpA leads to a pool of precursor in the cell wall which is available to repair openings in the S-layer formed during cell growth or following damage.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

