This is a preprint.
SARS-CoV-2 infection dynamics in lungs of African green monkeys
- PMID: 32839775
- PMCID: PMC7444286
- DOI: 10.1101/2020.08.20.258087
SARS-CoV-2 infection dynamics in lungs of African green monkeys
Update in
-
Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys.Sci Transl Med. 2021 Jan 27;13(578):eabe8146. doi: 10.1126/scitranslmed.abe8146. Epub 2021 Jan 11. Sci Transl Med. 2021. PMID: 33431511 Free PMC article.
Abstract
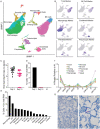
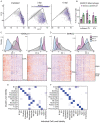
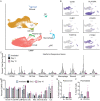
Detailed knowledge about the dynamics of SARS-CoV-2 infection is important for unraveling the viral and host factors that contribute to COVID-19 pathogenesis. Old-World nonhuman primates recapitulate mild-moderate COVID-19 cases, thereby serving as important pathogenesis models. We compared African green monkeys inoculated with SARS-CoV-2 or inactivated virus to study the dynamics of virus replication throughout the respiratory tract. RNA sequencing of single cells from the lungs and mediastinal lymph nodes allowed a high-resolution analysis of virus replication and host responses over time. Viral replication was mainly localized to the lower respiratory tract, with evidence of replication in the pneumocytes. Macrophages were found to play a role in initiating a pro-inflammatory state in the lungs, while also interacting with infected pneumocytes. Our dataset provides a detailed view of changes in host and virus replication dynamics over the course of mild COVID-19 and serves as a valuable resource to identify therapeutic targets.
Conflict of interest statement
Declaration of interests
The authors have no competing interests to declare.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
