Genetic examination of the marine bacterium Pseudoalteromonas luteoviolacea and effects of its metamorphosis-inducing factors
- PMID: 32840026
- PMCID: PMC8214333
- DOI: 10.1111/1462-2920.15211
Genetic examination of the marine bacterium Pseudoalteromonas luteoviolacea and effects of its metamorphosis-inducing factors
Abstract
Pseudoalteromonas luteoviolacea is a globally distributed marine bacterium that stimulates the metamorphosis of marine animal larvae, an important bacteria-animal interaction that can promote the recruitment of animals to benthic ecosystems. Recently, different P. luteoviolacea isolates have been shown to produce two stimulatory factors that can induce tubeworm and coral metamorphosis; Metamorphosis-Associated Contractile structures (MACs) and tetrabromopyrrole (TBP) respectively. However, it remains unclear what proportion of P. luteoviolacea isolates possess the genes encoding MACs, and what phenotypic effect MACs and TBP have on other larval species. Here, we show that 9 of 19 sequenced P. luteoviolacea genomes genetically encode both MACs and TBP. While P. luteoviolacea biofilms producing MACs stimulate the metamorphosis of the tubeworm Hydroides elegans, TBP biosynthesis genes had no effect under the conditions tested. Although MACs are lethal to larvae of the cnidarian Hydractinia symbiologicarpus, P. luteoviolacea mutants unable to produce MACs are capable of stimulating metamorphosis. Our findings reveal a hidden complexity of interactions between a single bacterial species, the factors it produces and two species of larvae belonging to different phyla.
© 2020 Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures


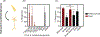
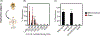
References
-
- Amador-Cano G, Carpizo-Ituarte E, and Cristino-Jorge D ( 2006) Role of protein kinase C, G-protein coupled receptors, and calcium flux during metamorphosis of the sea urchin Strongylocentrotus purpuratus. Biol Bull 210: 121–131. - PubMed
-
- Atencio LA, Dal Grande F, Young GO, Gavilán R, Guzmán HM, Schmitt I, et al. (2018) Antimicrobial-producing Pseudoalteromonas from the marine environment of Panama shows a high phylogenetic diversity and clonal structure. J Basic Microbiol 58: 747–769. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

