Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS, and Single-Molecule FRET
- PMID: 32840111
- PMCID: PMC9987321
- DOI: 10.1021/jacs.0c02088
Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS, and Single-Molecule FRET
Abstract
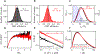
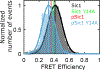
Intrinsically disordered proteins (IDPs) have fluctuating heterogeneous conformations, which makes their structural characterization challenging. Although challenging, characterization of the conformational ensembles of IDPs is of great interest, since their conformational ensembles are the link between their sequences and functions. An accurate description of IDP conformational ensembles depends crucially on the amount and quality of the experimental data, how it is integrated, and if it supports a consistent structural picture. We used integrative modeling and validation to apply conformational restraints and assess agreement with the most common structural techniques for IDPs: Nuclear Magnetic Resonance (NMR) spectroscopy, Small-angle X-ray Scattering (SAXS), and single-molecule Förster Resonance Energy Transfer (smFRET). Agreement with such a diverse set of experimental data suggests that details of the generated ensembles can now be examined with a high degree of confidence. Using the disordered N-terminal region of the Sic1 protein as a test case, we examined relationships between average global polymeric descriptions and higher-moments of their distributions. To resolve apparent discrepancies between smFRET and SAXS inferences, we integrated SAXS data with NMR data and reserved the smFRET data for independent validation. Consistency with smFRET, which was not guaranteed a priori, indicates that, globally, the perturbative effects of NMR or smFRET labels on the Sic1 ensemble are minimal. Analysis of the ensembles revealed distinguishing features of Sic1, such as overall compactness and large end-to-end distance fluctuations, which are consistent with biophysical models of Sic1's ultrasensitive binding to its partner Cdc4. Our results underscore the importance of integrative modeling and validation in generating and drawing conclusions from IDP conformational ensembles.
Conflict of interest statement
The authors declare no competing financial interests
Figures

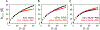


References
-
- Keith Dunker A, David Lawson J, Brown Celeste J, Williams Ryan M, Romero Pedro, Oh Jeong S, Oldfield Christopher J, Campen Andrew M, Ratliff Catherine M, Hipps Kerry W, Ausio Juan, Nissen Mark S, Reeves Raymond, Kang ChulHee, Kissinger Charles R, Bailey Robert W, Griswold Michael D, Chiu Wah, Garner Ethan C, and Obradovic Zoran. Intrinsically disordered protein. Journal of Molecular Graphics and Modelling, 19(1):26–59, February 2001. ISSN 1093-3263. doi: 10.1016/S1093-3263(00)00138-8. - DOI - PubMed
-
- Uversky Vladimir N., Gillespie Joel R., and Fink Anthony L.. Why Are “Natively Unfolded” Proteins Unstructured under Physiologic Conditions? Proteins: Structure, Function, and Bioinformatics, 41(3): 415–427, November 2000. ISSN 1097-0134. doi: 10.1002/1097-0134(20001115)41:3⟨415::AID-PROT13⟩3.0.CO;2-7. - DOI - PubMed
-
- Hofmann Hagen, Soranno Andrea, Borgia Alessandro, Gast Klaus, Nettels Daniel, and Schuler Benjamin. Polymer Scaling Laws of Unfolded and Intrinsically Disordered Proteins Quantified with Single-Molecule Spectroscopy. Proceedings of the National Academy of Sciences, 109(40):16155–16160, October 2012. ISSN 0027-8424, 1091-6490. doi: 10.1073/pnas.1207719109. - DOI - PMC - PubMed

