Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway
- PMID: 32840360
- PMCID: PMC7506943
- DOI: 10.1021/acschembio.0c00564
Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway
Abstract
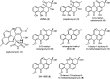
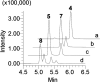
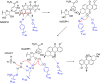
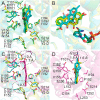
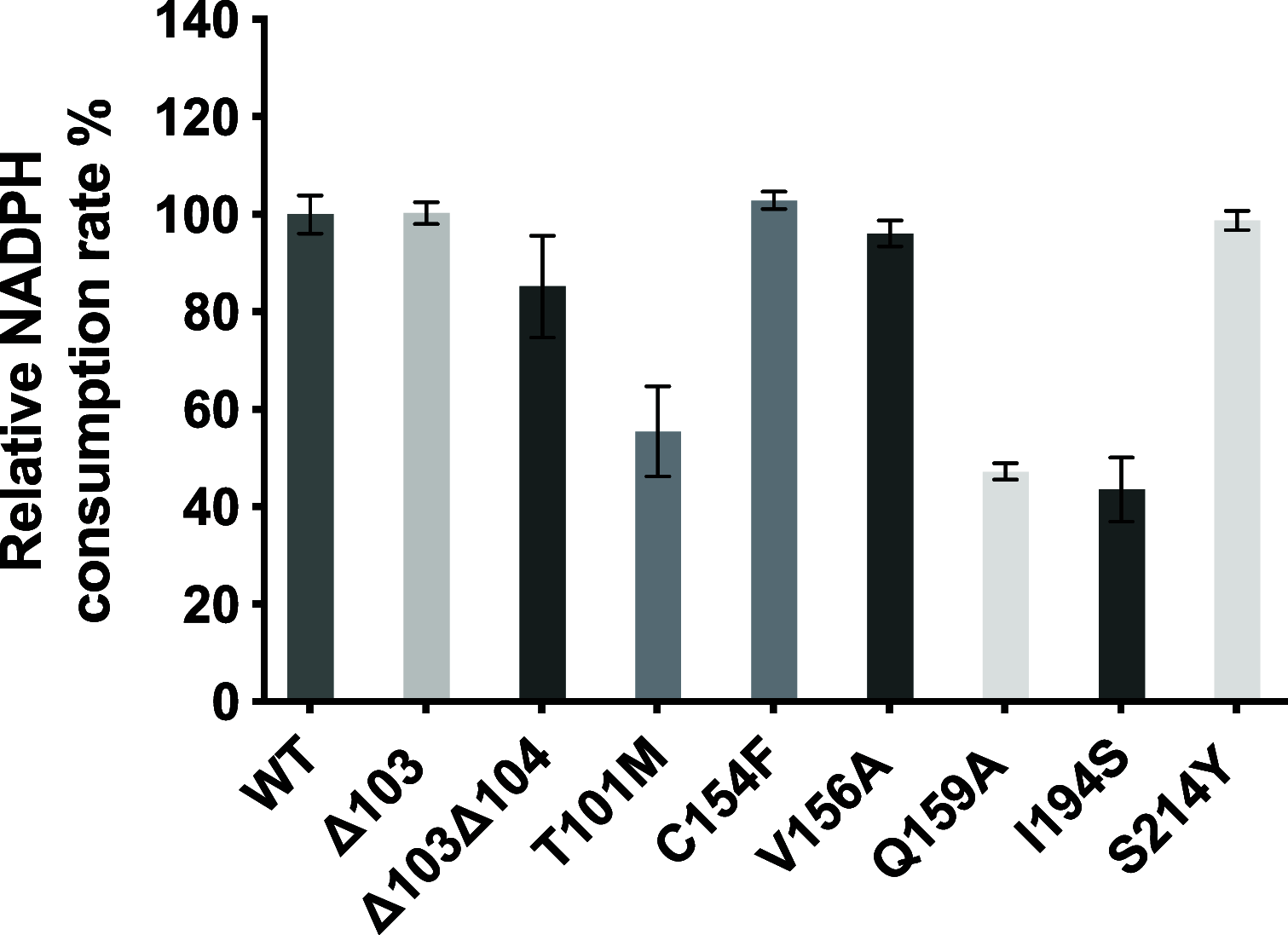
Angucyclines are a structurally diverse class of actinobacterial natural products defined by their varied polycyclic ring systems, which display a wide range of biological activities. We recently discovered lugdunomycin (1), a highly rearranged polyketide antibiotic derived from the angucycline backbone that is synthesized via several yet unexplained enzymatic reactions. Here, we show via in vivo, in vitro, and structural analysis that the promiscuous reductase LugOII catalyzes both a C6 and an unprecedented C1 ketoreduction. This then sets the stage for the subsequent C-ring cleavage that is key to the rearranged scaffolds of 1. The 1.1 Å structures of LugOII in complex with either ligand 8-O-Methylrabelomycin (4) or 8-O-Methyltetrangomycin (5) and of apoenzyme were resolved, which revealed a canonical Rossman fold and a remarkable conformational change during substrate capture and release. Mutational analysis uncovered key residues for substrate access, position, and catalysis as well as specific determinants that control its dual functionality. The insights obtained in this work hold promise for the discovery and engineering of other promiscuous reductases that may be harnessed for the generation of novel biocatalysts for chemoenzymatic applications.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

