Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy
- PMID: 32841044
- PMCID: PMC7676622
- DOI: 10.1161/CIRCGEN.120.002929
Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy
Abstract
Background: Pathogenic variants in MYBPC3, encoding cardiac MyBP-C (myosin binding protein C), are the most common cause of familial hypertrophic cardiomyopathy. A large number of unique MYBPC3 variants and relatively small genotyped hypertrophic cardiomyopathy cohorts have precluded detailed genotype-phenotype correlations.
Methods: Patients with hypertrophic cardiomyopathy and MYBPC3 variants were identified from the Sarcomeric Human Cardiomyopathy Registry. Variant types and locations were analyzed, morphological severity was assessed, and time-event analysis was performed (composite clinical outcome of sudden death, class III/IV heart failure, left ventricular assist device/transplant, atrial fibrillation). For selected missense variants falling in enriched domains, myofilament localization and degradation rates were measured in vitro.
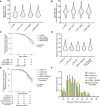
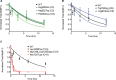
Results: Among 4756 genotyped patients with hypertrophic cardiomyopathy in Sarcomeric Human Cardiomyopathy Registry, 1316 patients were identified with adjudicated pathogenic truncating (N=234 unique variants, 1047 patients) or nontruncating (N=22 unique variants, 191 patients) variants in MYBPC3. Truncating variants were evenly dispersed throughout the gene, and hypertrophy severity and outcomes were not associated with variant location (grouped by 5'-3' quartiles or by founder variant subgroup). Nontruncating pathogenic variants clustered in the C3, C6, and C10 domains (18 of 22, 82%, P<0.001 versus Genome Aggregation Database common variants) and were associated with similar hypertrophy severity and adverse event rates as observed with truncating variants. MyBP-C with variants in the C3, C6, and C10 domains was expressed in rat ventricular myocytes. C10 mutant MyBP-C failed to incorporate into myofilaments and degradation rates were accelerated by ≈90%, while C3 and C6 mutant MyBP-C incorporated normally with degradation rate similar to wild-type.
Conclusions: Truncating variants account for 91% of MYBPC3 pathogenic variants and cause similar clinical severity and outcomes regardless of location, consistent with locus-independent loss-of-function. Nontruncating MYBPC3 pathogenic variants are regionally clustered, and a subset also cause loss of function through failure of myofilament incorporation and rapid degradation. Cardiac morphology and clinical outcomes are similar in patients with truncating versus nontruncating variants.
Keywords: actins; genotype; hypertrophic cardiomyopathy; myosin; sarcomere.
Figures



References
-
- Ho CY, Day SM, Ashley EA, Michels M, Pereira AC, Jacoby D, Cirino AL, Fox JC, Lakdawala NK, Ware JS, et al. Genotype and lifetime burden of disease in hypertrophic cardiomyopathy: insights from the sarcomeric human cardiomyopathy registry (SHaRe). Circulation. 2018;138:1387–1398. doi: 10.1161/CIRCULATIONAHA.117.033200 - PMC - PubMed
-
- Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ, Ackerman MJ. Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:1903–1910. doi: 10.1016/j.jacc.2004.07.045 - PubMed
-
- Alfares AA, Kelly MA, McDermott G, Funke BH, Lebo MS, Baxter SB, Shen J, McLaughlin HM, Clark EH, Babb LJ, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–8. doi: 10.1038/gim.2014.205 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

