Glycan cross-feeding supports mutualism between Fusobacterium and the vaginal microbiota
- PMID: 32841232
- PMCID: PMC7447053
- DOI: 10.1371/journal.pbio.3000788
Glycan cross-feeding supports mutualism between Fusobacterium and the vaginal microbiota
Abstract
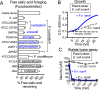
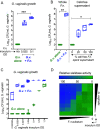
Women with bacterial vaginosis (BV), an imbalance of the vaginal microbiome, are more likely to be colonized by potential pathogens such as Fusobacterium nucleatum, a bacterium linked with intrauterine infection and preterm birth. However, the conditions and mechanisms supporting pathogen colonization during vaginal dysbiosis remain obscure. We demonstrate that sialidase activity, a diagnostic feature of BV, promoted F. nucleatum foraging and growth on mammalian sialoglycans, a nutrient resource that was otherwise inaccessible because of the lack of endogenous F. nucleatum sialidase. In mice with sialidase-producing vaginal microbiotas, mutant F. nucleatum unable to consume sialic acids was impaired in vaginal colonization. These experiments in mice also led to the discovery that F. nucleatum may also "give back" to the community by reinforcing sialidase activity, a biochemical feature of human dysbiosis. Using human vaginal bacterial communities, we show that F. nucleatum supported robust outgrowth of Gardnerella vaginalis, a major sialidase producer and one of the most abundant organisms in BV. These results illustrate that mutually beneficial relationships between vaginal bacteria support pathogen colonization and may help maintain features of dysbiosis. These findings challenge the simplistic dogma that the mere absence of "healthy" lactobacilli is the sole mechanism that creates a permissive environment for pathogens during vaginal dysbiosis. Given the ubiquity of F. nucleatum in the human mouth, these studies also suggest a possible mechanism underlying links between vaginal dysbiosis and oral sex.
Conflict of interest statement
The authors declare that they have no relevant conflicts of interest.
Figures






References
-
- Hill GB, Eschenbach DA, Holmes KK. Bacteriology of the vagina. Scand J Urol Nephrol Suppl. 1984;86:23–39. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

