Comparative study on three viral enrichment approaches based on RNA extraction for plant virus/viroid detection using high-throughput sequencing
- PMID: 32841302
- PMCID: PMC7447037
- DOI: 10.1371/journal.pone.0237951
Comparative study on three viral enrichment approaches based on RNA extraction for plant virus/viroid detection using high-throughput sequencing
Abstract
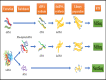
High-throughput sequencing (HTS) has become increasingly popular as virus diagnostic tool. It has been used to detect and identify plant viruses and viroids in different types of matrices and tissues. A viral sequence enrichment method prior to HTS is required to increase the viral reads in the generated data to ease the bioinformatic analysis of generated sequences. In this study, we compared the sensitivity of three viral enrichment approaches, i.e. double stranded RNA (dsRNA), ribosomal RNA depleted total RNA (ribo-depleted totRNA) and small RNA (sRNA) for plant virus/viroid detection, followed by sequencing on MiSeq and NextSeq Illumina platforms. The three viral enrichment approaches used here enabled the detection of all viruses/viroid used in this study. When the data was normalised, the recovered viral/viroid nucleotides and depths were depending on the viral genome and the enrichment method used. Both dsRNA and ribo-depleted totRNA approaches detected a divergent strain of Wuhan aphid virus 2 that was not expected in this sample. Additionally, Vicia cryptic virus was detected in the data of dsRNA and sRNA approaches only. The results suggest that dsRNA enrichment has the highest potential to detect and identify plant viruses and viroids. The dsRNA approach used here detected all viruses/viroid, consumed less time, was lower in cost, and required less starting material. Therefore, this approach appears to be suitable for diagnostics laboratories.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures



References
-
- Canto T, Aranda MA, Fereres A (2009) Climate change effects on physiology and population processes of hosts and vectors that influence the spread of hemipteran-borne plant viruses. Glob Change Biol 15: 1884–1894.
-
- Jones RAC (2016) Future scenarios for plant virus pathogens as climate change progresses. Adv Vir Res 95: 87–147. - PubMed
-
- Myers SS, Smith MR, Guth S, Golden CD, Vaitla B et al. (2017) Climate change and global food systems: potential impacts on food security and undernutrition. Ann Rev Public Health 38: 259–277. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

